Protein length in eukaryotic and prokaryotic proteomes
- PMID: 15951512
- PMCID: PMC1150220
- DOI: 10.1093/nar/gki615
Protein length in eukaryotic and prokaryotic proteomes
Abstract
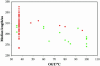
We analyzed length differences of eukaryotic, bacterial and archaeal proteins in relation to function, conservation and environmental factors. Comparing Eukaryotes and Prokaryotes, we found that the greater length of eukaryotic proteins is pervasive over all functional categories and involves the vast majority of protein families. The magnitude of these differences suggests that the evolution of eukaryotic proteins was influenced by processes of fusion of single-function proteins into extended multi-functional and multi-domain proteins. Comparing Bacteria and Archaea, we determined that the small but significant length difference observed between their proteins results from a combination of three factors: (i) bacterial proteomes include a greater proportion than archaeal proteomes of longer proteins involved in metabolism or cellular processes, (ii) within most functional classes, protein families unique to Bacteria are generally longer than protein families unique to Archaea and (iii) within the same protein family, homologs from Bacteria tend to be longer than the corresponding homologs from Archaea. These differences are interpreted with respect to evolutionary trends and prevailing environmental conditions within the two prokaryotic groups.
Figures
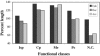

References
-
- Galperin M.Y., Tatusov R.L., Koonin E.V. In: Organization of the Prokaryotic Genome. Charlebois R.L., editor. Washington, DC: ASM Press; 1999.
-
- Zhang J. Protein-length distributions for the three domains of life. Trends Genet. 2000;16:107–109. - PubMed
-
- Liang P., Riley M. A comparative genomics approach for studying ancestral proteins and evolution. Adv. Appl. Microbiol. 2001;50:39–72. - PubMed
-
- Skovgaard M., Jensen L.J., Brunak S., Ussery D., Krogh A. On the total number of genes and their length distribution in complete microbial genomes. Trends Genet. 2001;17:425–428. - PubMed
-
- Karlin S., Brocchieri L., Trent J., Blaisdell B.E., Mrazek J. Heterogeneity of genome and proteome content in bacteria, archaea, and eukaryotes. Theor. Popul. Biol. 2002;61:367–390. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

