Multiplexed detection of pathogen DNA with DNA-based fluorescence nanobarcodes
- PMID: 15951805
- PMCID: PMC7097058
- DOI: 10.1038/nbt1106
Multiplexed detection of pathogen DNA with DNA-based fluorescence nanobarcodes
Abstract
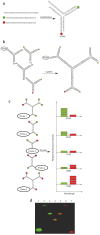
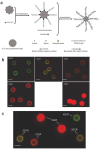
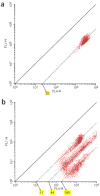
Rapid, multiplexed, sensitive and specific molecular detection is of great demand in gene profiling, drug screening, clinical diagnostics and environmental analysis. One of the major challenges in multiplexed analysis is to identify each specific reaction with a distinct label or 'code'. Two encoding strategies are currently used: positional encoding, in which every potential reaction is preassigned a particular position on a solid-phase support such as a DNA microarray, and reaction encoding, where every possible reaction is uniquely tagged with a code that is most often optical or particle based. The micrometer size, polydispersity, complex fabrication process and nonbiocompatibility of current codes limit their usability. Here we demonstrate the synthesis of dendrimer-like DNA-based, fluorescence-intensity-coded nanobarcodes, which contain a built-in code and a probe for molecular recognition. Their application to multiplexed detection of the DNA of several pathogens is first shown using fluorescence microscopy and dot blotting, and further demonstrated using flow cytometry that resulted in detection that was sensitive (attomole) and rapid.
Conflict of interest statement
A patent for similar technology is being filed, the value of which may be increased by this publication.
Figures

References
-
- Fulton RJ, McDade RL, Smith PL, Kienker LJ, Kettman JR. Advanced multiplexed analysis with the FlowMetrix(TM) system. Clin. Chem. 1997;43:1749–1756. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

