RNA interference is ineffective as a routine method for gene silencing in chick embryos as monitored by fgf8 silencing
- PMID: 15951844
- PMCID: PMC1140352
- DOI: 10.7150/ijbs.1.1
RNA interference is ineffective as a routine method for gene silencing in chick embryos as monitored by fgf8 silencing
Abstract
The in vivo accessibility of the chick embryo makes it a favoured model system for experimental developmental biology. Although the range of available techniques now extends to miss-expression of genes through in ovo electroporation, it remains difficult to knock out individual gene expression. Recently, the possibility of silencing gene expression by RNAi in chick embryos has been reported. However, published studies show only discrete quantitative differences in the expression of the endogenous targeted genes and unclear morphological alterations. To elucidate whether the tools currently available are adequate to silence gene expression sufficiently to produce a clear and specific null-like mutant phenotype, we have performed several experiments with different molecules that trigger RNAi: dsRNA, siRNA, and shRNA produced from a plasmid coexpressing green fluorescent protein as an internal marker. Focussing on fgf8 expression in the developing isthmus, we show that no morphological defects are observed, and that fgf8 expression is neither silenced in embryos microinjected with dsRNA nor in embryos microinjected and electroporated with a pool of siRNAs. Moreover, fgf8 expression was not significantly silenced in most isthmic cells transformed with a plasmid producing engineered shRNAs to fgf8. We also show that siRNA molecules do not spread significantly from cell to cell as reported for invertebrates, suggesting the existence of molecular differences between different model systems that may explain the different responses to RNAi. Although our results are basically in agreement with previously reported studies, we suggest, in contrast to them, that with currently available tools and techniques the number of cells in which fgf8 gene expression is decreased, if any, is not sufficient to generate a detectable mutant phenotype, thus making RNAi useless as a routine method for functional gene analysis in chick embryos.
Conflict of interest statement
Figures
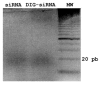






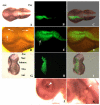
References
-
- Muramatsu T, Mizutani Y, Ohmori Y, Okumura J. Comparison of three nonviral transfection methods for foreign gene expression in early chicken embryos in ovo. Biochem Biophys Res Commun. 1997;230:376–380. - PubMed
-
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. - PubMed
-
- Bass BL. Double-stranded RNA as a template for gene silencing. Cell. 2000;101:235–238. - PubMed
-
- Bueno D, Romero R, Hernández Hernández V, Sancho V, Fernández-Rodríguez J, Cardona AT, Vila-Farré M. RNA interference: a new and powerful tool for functional genomic analysis. C Sci. 2001;2:23–33.
-
- Hanon GJ. RNA interference. Nature. 2002;418:244–250. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

