Identification of the hydrophobic strand in the A-B loop of leptin as major binding site III: implications for large-scale preparation of potent recombinant human and ovine leptin antagonists
- PMID: 15952938
- PMCID: PMC1276919
- DOI: 10.1042/BJ20050457
Identification of the hydrophobic strand in the A-B loop of leptin as major binding site III: implications for large-scale preparation of potent recombinant human and ovine leptin antagonists
Abstract
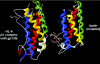
Interaction of leptin with its receptors resembles that of interleukin-6 and granulocyte colony-stimulating factor, which interact with their receptors through binding sites I-III. Site III plays a pivotal role in receptors' dimerization or tetramerization and subsequent activation. Leptin's site III also mediates the formation of an active multimeric complex through its interaction with the IGD (immunoglobulin-like domain) of LEPRs (leptin receptors). Using a sensitive hydrophobic cluster analysis of leptin's and LEPR's sequences, we identified hydrophobic stretches in leptin's A-B loop (amino acids 39-42) and in the N-terminal end of LEPR's IGD (amino acids 325-328) that are predicted to participate in site III and to interact with each other in a beta-sheet-like configuration. To verify this hypothesis, we prepared and purified to homogeneity (as verified by SDS/PAGE, gel filtration and reverse-phase chromatography) several alanine muteins of amino acids 39-42 in human and ovine leptins. CD analyses revealed that those mutations hardly affect the secondary structure. All muteins acted as true antagonists, i.e. they bound LEPR with an affinity similar to the wild-type hormone, had no agonistic activity and specifically inhibited leptin action in several leptin-responsive in vitro bioassays. Alanine mutagenesis of LEPR's IGD (amino acids 325-328) drastically reduced its biological but not binding activity, indicating the importance of this region for interaction with leptin's site III. FRET (fluorescence resonance energy transfer) microscopy experiments have documented that the transient FRET signalling occurring upon exposure to leptin results not from binding of the ligand, but from ligand-induced oligomerization of LEPRs mediated by leptin's site III.
Figures







References
-
- Zhang Y., Proenca R., Maffei M., Barone M., Leopold L., Friedman J. M. Positional cloning of the mouse obese gene and its human homologue. Nature (London) 1994;372:425–432. - PubMed
-
- Tartaglia L. A., Dembski M., Weng X., Deng N., Culpepper J., Devos R., Richards G. J., Campfield L. A., Clark F. T., Deeds J., et al. Identification and expression cloning of a leptin receptor, OB-R. Cell (Cambridge, Mass.) 1995;83:1263–1271. - PubMed
-
- Zhang F., Basinski M. B., Beals J. M., Briggs S. L., Churgay L. M., Clawson D. K., DiMarchi R. D., Furman T. C., Hale J. E., Hsiung H. M., et al. Crystal structure of the obese protein leptin-E100. Nature (London) 1997;387:206–209. - PubMed
-
- Zabeau L., Defeau D., Van der Heyden J., Iserentant H., Vandekerckhove J., Tavernier J. The ins and outs of leptin receptor activation. FEBS Lett. 2003;18:150–161. - PubMed
-
- Fong T. M., Huang R. R., Tota M. R., Mao C., Smith T., Varnerin J., Karpitskiy V. V., Krause J. E., Van der Ploeg L. H. Localization of leptin binding domain in the leptin receptor. Mol. Pharmacol. 1998;53:234–240. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

