Characterization of a thermostable UvrD helicase and its participation in helicase-dependent amplification
- PMID: 15955821
- PMCID: PMC1361353
- DOI: 10.1074/jbc.M503096200
Characterization of a thermostable UvrD helicase and its participation in helicase-dependent amplification
Abstract
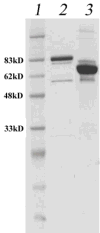
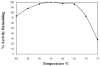
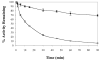
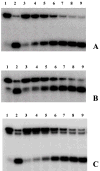
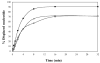
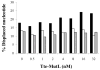
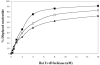
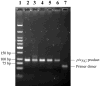
Helicase-dependent amplification (HDA) is an isothermal in vitro DNA amplification method based upon the coordinated actions of helicases to separate double-stranded DNA and DNA polymerases to synthesize DNA. Previously, a mesophilic form of HDA (mHDA) utilizing the Escherichia coli UvrD helicase, DNA polymerase I Klenow fragment, two accessory proteins, MutL and single-stranded DNA-binding protein (SSB), was developed (1). In an effort to improve the specificity and performance of HDA, we have cloned and purified a thermostable UvrD helicase (Tte-UvrD) and the mutL homolog (Tte-MutL) from Thermoanaerobacter tengcongensis. Characterization of the Tte-UvrD helicase shows that it is stable and active from 45 to 65 degrees C. We have found that the Tte-UvrD helicase unwinds blunt-ended DNA duplexes as well as substrates possessing 3'- or 5'-ssDNA tails. Tte-UvrD was used to develop athermophilichelicase-dependent amplification (tHDA) system to selectively amplify target sequences at 60-65 degrees C. The tHDA system is more efficient than mHDA, displaying heightened amplification sensitivity without the need for the MutL and SSB accessory proteins. Amplification independent of MutL corresponds with studies demonstrating that maximal Tte-UvrD helicase activity does not require the mutL homolog. The tHDA system allows for rapid amplification and detection of targets present in genomic DNA. The expeditious nature and simplistic design of the tHDA platform makes the technology ideal for use in diagnostic applications requiring rapid identification of organisms at the point-of-need.
Figures



Similar articles
-
A human PMS2 homologue from Aquifex aeolicus stimulates an ATP-dependent DNA helicase.J Biol Chem. 2010 Apr 9;285(15):11087-92. doi: 10.1074/jbc.M109.050955. Epub 2010 Feb 2. J Biol Chem. 2010. PMID: 20129926 Free PMC article.
-
A Dimer of Escherichia coli UvrD is the active form of the helicase in vitro.J Mol Biol. 2003 Jan 31;325(5):913-35. doi: 10.1016/s0022-2836(02)01277-9. J Mol Biol. 2003. PMID: 12527299
-
An oligomeric form of E. coli UvrD is required for optimal helicase activity.J Mol Biol. 1999 Nov 5;293(4):815-34. doi: 10.1006/jmbi.1999.3185. J Mol Biol. 1999. PMID: 10543970
-
Isothermal DNA amplification in vitro: the helicase-dependent amplification system.Cell Mol Life Sci. 2009 Oct;66(20):3325-36. doi: 10.1007/s00018-009-0094-3. Epub 2009 Jul 24. Cell Mol Life Sci. 2009. PMID: 19629390 Free PMC article. Review.
-
Roles of the C-Terminal Amino Acids of Non-Hexameric Helicases: Insights from Escherichia coli UvrD.Int J Mol Sci. 2021 Jan 20;22(3):1018. doi: 10.3390/ijms22031018. Int J Mol Sci. 2021. PMID: 33498436 Free PMC article. Review.
Cited by
-
Virus Detection: A Review of the Current and Emerging Molecular and Immunological Methods.Front Mol Biosci. 2021 Apr 20;8:637559. doi: 10.3389/fmolb.2021.637559. eCollection 2021. Front Mol Biosci. 2021. PMID: 33959631 Free PMC article. Review.
-
Alteration of enzymes and their application to nucleic acid amplification (Review).Int J Mol Med. 2020 Nov;46(5):1633-1643. doi: 10.3892/ijmm.2020.4726. Epub 2020 Sep 15. Int J Mol Med. 2020. PMID: 33000189 Free PMC article. Review.
-
Developments in integrating nucleic acid isothermal amplification and detection systems for point-of-care diagnostics.Biosens Bioelectron. 2020 Dec 15;170:112674. doi: 10.1016/j.bios.2020.112674. Epub 2020 Oct 2. Biosens Bioelectron. 2020. PMID: 33035900 Free PMC article. Review.
-
Development and comparison of a rapid isothermal nucleic acid amplification test for typing of herpes simplex virus types 1 and 2 on a portable fluorescence detector.J Mol Diagn. 2012 Nov;14(6):569-76. doi: 10.1016/j.jmoldx.2012.05.005. Epub 2012 Aug 27. J Mol Diagn. 2012. PMID: 22951487 Free PMC article.
-
Intravesicle isothermal DNA replication.BMC Res Notes. 2011 Apr 15;4:128. doi: 10.1186/1756-0500-4-128. BMC Res Notes. 2011. PMID: 21496266 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

