An electrostatic engine model for autoinhibition and activation of the epidermal growth factor receptor (EGFR/ErbB) family
- PMID: 15955874
- PMCID: PMC2266615
- DOI: 10.1085/jgp.200509274
An electrostatic engine model for autoinhibition and activation of the epidermal growth factor receptor (EGFR/ErbB) family
Abstract
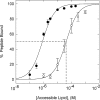
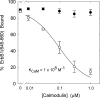
We propose a new mechanism to explain autoinhibition of the epidermal growth factor receptor (EGFR/ErbB) family of receptor tyrosine kinases based on a structural model that postulates both their juxtamembrane and protein tyrosine kinase domains bind electrostatically to acidic lipids in the plasma membrane, restricting access of the kinase domain to substrate tyrosines. Ligand-induced dimerization promotes partial trans autophosphorylation of ErbB1, leading to a rapid rise in intracellular [Ca(2+)] that can activate calmodulin. We postulate the Ca(2+)/calmodulin complex binds rapidly to residues 645--660 of the juxtamembrane domain, reversing its net charge from +8 to -8 and repelling it from the negatively charged inner leaflet of the membrane. The repulsion has two consequences: it releases electrostatically sequestered phosphatidylinositol 4,5-bisphosphate (PIP(2)), and it disengages the kinase domain from the membrane, allowing it to become fully active and phosphorylate an adjacent ErbB molecule or other substrate. We tested various aspects of the model by measuring ErbB juxtamembrane peptide binding to phospholipid vesicles using both a centrifugation assay and fluorescence correlation spectroscopy; analyzing the kinetics of interactions between ErbB peptides, membranes, and Ca(2+)/calmodulin using fluorescence stop flow; assessing ErbB1 activation in Cos1 cells; measuring fluorescence resonance energy transfer between ErbB peptides and PIP(2); and making theoretical electrostatic calculations on atomic models of membranes and ErbB juxtamembrane and kinase domains.
Figures



Similar articles
-
Electrostatic interactions of peptides flanking the tyrosine kinase domain in the epidermal growth factor receptor provides a model for intracellular dimerization and autophosphorylation.Proteins. 2006 Mar 1;62(4):1036-43. doi: 10.1002/prot.20780. Proteins. 2006. PMID: 16380971
-
EGFR juxtamembrane domain, membranes, and calmodulin: kinetics of their interaction.Biophys J. 2009 Jun 17;96(12):4887-95. doi: 10.1016/j.bpj.2009.03.027. Biophys J. 2009. PMID: 19527647 Free PMC article.
-
Membrane-permeable calmodulin inhibitors (e.g. W-7/W-13) bind to membranes, changing the electrostatic surface potential: dual effect of W-13 on epidermal growth factor receptor activation.J Biol Chem. 2007 Mar 16;282(11):8474-86. doi: 10.1074/jbc.M607211200. Epub 2007 Jan 16. J Biol Chem. 2007. PMID: 17227773
-
The ErbB/HER family of protein-tyrosine kinases and cancer.Pharmacol Res. 2014 Jan;79:34-74. doi: 10.1016/j.phrs.2013.11.002. Epub 2013 Nov 20. Pharmacol Res. 2014. PMID: 24269963 Review.
-
The ErbB/HER receptor protein-tyrosine kinases and cancer.Biochem Biophys Res Commun. 2004 Jun 18;319(1):1-11. doi: 10.1016/j.bbrc.2004.04.150. Biochem Biophys Res Commun. 2004. PMID: 15158434 Review.
Cited by
-
From Orai to E-Cadherin: Subversion of Calcium Trafficking in Cancer to Drive Proliferation, Anoikis-Resistance, and Metastasis.Biomedicines. 2020 Jun 21;8(6):169. doi: 10.3390/biomedicines8060169. Biomedicines. 2020. PMID: 32575848 Free PMC article. Review.
-
Epidermal growth factor receptor activation remodels the plasma membrane lipid environment to induce nanocluster formation.Mol Cell Biol. 2010 Aug;30(15):3795-804. doi: 10.1128/MCB.01615-09. Epub 2010 Jun 1. Mol Cell Biol. 2010. PMID: 20516214 Free PMC article.
-
The EGFR family: not so prototypical receptor tyrosine kinases.Cold Spring Harb Perspect Biol. 2014 Apr 1;6(4):a020768. doi: 10.1101/cshperspect.a020768. Cold Spring Harb Perspect Biol. 2014. PMID: 24691965 Free PMC article. Review.
-
Putting together structures of epidermal growth factor receptors.Curr Opin Struct Biol. 2014 Dec;29:95-101. doi: 10.1016/j.sbi.2014.10.002. Epub 2014 Oct 24. Curr Opin Struct Biol. 2014. PMID: 25460273 Free PMC article. Review.
-
Regulation of EGFR nanocluster formation by ionic protein-lipid interaction.Cell Res. 2014 Aug;24(8):959-76. doi: 10.1038/cr.2014.89. Epub 2014 Jul 8. Cell Res. 2014. PMID: 25001389 Free PMC article.
References
-
- Aifa, S., K. Johansen, U.K. Nilsson, B. Liedberg, I. Lundstrom, and S.P.S. Svensson. 2002. Interactions between the juxtamembrane domain of the EGFR and calmodulin measured by surface plasmon resonance. Cell. Signal. 14:1005–1013. - PubMed
-
- Aifa, S., J. Aydin, G. Nordvall, I. Lundstrom, S.P.S. Svensson, and O. Hermanson. 2005. A basic peptide within the juxtamembrane region is required for EGF receptor dimerization. Exp. Cell Res. 302:108–114. - PubMed
-
- Arbuzova, A., J. Wang, D. Murray, J. Jacob, D.S. Cafiso, and S. McLaughlin. 1997. Kinetics of interaction of the myristoylated alanine-rich C kinase substrate, membranes, and calmodulin. J. Biol. Chem. 272:27167–27177. - PubMed
-
- Arbuzova, A., D. Murray, and S. McLaughlin. 1998. MARCKS, membranes and calmodulin: kinetics of their interaction. Biochim. Biophys. Acta. 1376:369–379. - PubMed
-
- Arbuzova, A., L. Wang, J. Wang, G. Hangyàs-Mihàlynè, D. Murray, B. Honig, and S. McLaughlin. 2000. Membrane binding of peptides containing both basic and aromatic residues. Biochemistry. 39:10330–10339. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

