Phylogenomic analysis of the receptor-like proteins of rice and Arabidopsis
- PMID: 15955925
- PMCID: PMC1150382
- DOI: 10.1104/pp.104.054452
Phylogenomic analysis of the receptor-like proteins of rice and Arabidopsis
Abstract
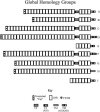
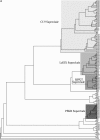
The tomato (Lycopersicon esculentum) Cf-9 resistance gene encodes the first characterized member of the plant receptor-like protein (RLP) family. Other RLPs such as CLAVATA2 and TOO MANY MOUTHS are known to regulate development. The domain structure of RLPs consists of extracellular leucine-rich repeats, a transmembrane helix, and a short cytoplasmic region. Here, we identify 90 RLPs in rice (Oryza sativa) and compare them with functionally characterized RLPs from different plant species and with 56 Arabidopsis (Arabidopsis thaliana) RLPs, including the downy mildew resistance protein RPP27. Many RLPs cluster into four distinct superclades, three of which include RLPs known to be involved in plant defense. Sequence comparisons reveal diagnostic amino acid residues that may specify different molecular functions in different RLP subtypes. This analysis of rice RLPs thus identified at least 73 candidate resistance genes and four genes potentially involved in development. Due to the synteny between rice and other Gramineae, this analysis should provide valuable tools for experimental studies in rice and other cereals.
Figures





Similar articles
-
Genome cluster database. A sequence family analysis platform for Arabidopsis and rice.Plant Physiol. 2005 May;138(1):47-54. doi: 10.1104/pp.104.059048. Plant Physiol. 2005. PMID: 15888677 Free PMC article.
-
Arabidopsis downy mildew resistance gene RPP27 encodes a receptor-like protein similar to CLAVATA2 and tomato Cf-9.Plant Physiol. 2004 Jun;135(2):1100-12. doi: 10.1104/pp.103.037770. Epub 2004 May 21. Plant Physiol. 2004. Retraction in: Plant Physiol. 2007 Feb;143(2):1079. doi: 10.1104/pp.104.900215. PMID: 15155873 Free PMC article. Retracted.
-
Genome-wide analysis of heat shock transcription factor families in rice and Arabidopsis.J Genet Genomics. 2008 Feb;35(2):105-18. doi: 10.1016/S1673-8527(08)60016-8. J Genet Genomics. 2008. PMID: 18407058
-
The plant B3 superfamily.Trends Plant Sci. 2008 Dec;13(12):647-55. doi: 10.1016/j.tplants.2008.09.006. Epub 2008 Nov 3. Trends Plant Sci. 2008. PMID: 18986826 Review.
-
[Plant genome sequencing: a prelude to the study of its expression].J Soc Biol. 2002;196(4):297-301. J Soc Biol. 2002. PMID: 12645299 Review. French.
Cited by
-
Ectopic expression of a novel Ser/Thr protein kinase from cotton (Gossypium barbadense), enhances resistance to Verticillium dahliae infection and oxidative stress in Arabidopsis.Plant Cell Rep. 2013 Nov;32(11):1703-13. doi: 10.1007/s00299-013-1481-7. Epub 2013 Aug 4. Plant Cell Rep. 2013. PMID: 23912851
-
Crystal structure of the extracellular domain of the receptor-like kinase TMK3 from Arabidopsis thaliana.Acta Crystallogr F Struct Biol Commun. 2020 Aug 1;76(Pt 8):384-390. doi: 10.1107/S2053230X20010122. Epub 2020 Jul 29. Acta Crystallogr F Struct Biol Commun. 2020. PMID: 32744250 Free PMC article.
-
A malectin-like/leucine-rich repeat receptor protein kinase gene, RLK-V, regulates powdery mildew resistance in wheat.Mol Plant Pathol. 2018 Dec;19(12):2561-2574. doi: 10.1111/mpp.12729. Epub 2018 Oct 16. Mol Plant Pathol. 2018. PMID: 30030900 Free PMC article.
-
Genome analysis of poplar LRR-RLP gene clusters reveals RISP, a defense-related gene coding a candidate endogenous peptide elicitor.Front Plant Sci. 2014 Mar 28;5:111. doi: 10.3389/fpls.2014.00111. eCollection 2014. Front Plant Sci. 2014. PMID: 24734035 Free PMC article.
-
Genetic dissection of Verticillium wilt resistance mediated by tomato Ve1.Plant Physiol. 2009 May;150(1):320-32. doi: 10.1104/pp.109.136762. Epub 2009 Mar 25. Plant Physiol. 2009. PMID: 19321708 Free PMC article.
References
-
- Bonifacino JS, Traub LM (2003) Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annu Rev Biochem 72: 395–447 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

