Structure of RadB recombinase from a hyperthermophilic archaeon, Thermococcus kodakaraensis KOD1: an implication for the formation of a near-7-fold helical assembly
- PMID: 15956102
- PMCID: PMC1150280
- DOI: 10.1093/nar/gki662
Structure of RadB recombinase from a hyperthermophilic archaeon, Thermococcus kodakaraensis KOD1: an implication for the formation of a near-7-fold helical assembly
Abstract
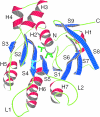
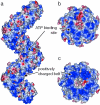
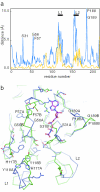
The X-ray crystal structure of RadB from Thermococcus kodakaraensis KOD1, an archaeal homologue of the RecA/Rad51 family proteins, have been determined in two crystal forms. The structure represents the core ATPase domain of the RecA/Rad51 proteins. Two independent molecules in the type 1 crystal were roughly related by 7-fold screw symmetry whereas non-crystallographic 2-fold symmetry was observed in the type 2 crystal. The dimer structure in the type 1 crystal is extended to construct a helical assembly, which resembles the filamentous structures reported for other RecA/Rad51 proteins. The molecular interface in the type 1 dimer is formed by facing a basic surface patch of one monomer to an acidic one of the other. The empty ATP binding pocket is located at the interface and barely concealed from the outside similarly to that in the active form of the RecA filament. The model assembly has a positively charged belt on one surface bordering the helical groove suitable for facile binding of DNA. Electron microscopy has revealed that, in the absence of ATP and DNA, RadB forms a filament with a similar diameter to that of the hypothetical assembly, although its helical properties were not confirmed.
Figures




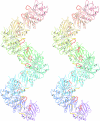

Similar articles
-
Interactions of RadB, a DNA repair protein in archaea, with DNA and ATP.J Mol Biol. 2006 Apr 21;358(1):46-56. doi: 10.1016/j.jmb.2006.02.010. Epub 2006 Feb 21. J Mol Biol. 2006. PMID: 16516228
-
Crystal structure of the left-handed archaeal RadA helical filament: identification of a functional motif for controlling quaternary structures and enzymatic functions of RecA family proteins.Nucleic Acids Res. 2007;35(6):1787-801. doi: 10.1093/nar/gkl1131. Epub 2007 Feb 28. Nucleic Acids Res. 2007. PMID: 17329376 Free PMC article.
-
Crystal structure of phosphopantothenate synthetase from Thermococcus kodakarensis.Proteins. 2014 Sep;82(9):1924-36. doi: 10.1002/prot.24546. Epub 2014 Apr 16. Proteins. 2014. PMID: 24638914
-
Crystal structure of highly thermostable glycerol kinase from a hyperthermophilic archaeon in a dimeric form.FEBS J. 2008 May;275(10):2632-43. doi: 10.1111/j.1742-4658.2008.06410.x. Epub 2008 Apr 17. FEBS J. 2008. PMID: 18422647
-
What is the structure of the RecA-DNA filament?Curr Protein Pept Sci. 2004 Apr;5(2):73-9. doi: 10.2174/1389203043486883. Curr Protein Pept Sci. 2004. PMID: 15078218 Review.
Cited by
-
An overview of 25 years of research on Thermococcus kodakarensis, a genetically versatile model organism for archaeal research.Folia Microbiol (Praha). 2020 Feb;65(1):67-78. doi: 10.1007/s12223-019-00730-2. Epub 2019 Jul 8. Folia Microbiol (Praha). 2020. PMID: 31286382 Review.
-
Role of allosteric switch residue histidine 195 in maintaining active-site asymmetry in presynaptic filaments of bacteriophage T4 UvsX recombinase.J Mol Biol. 2009 Jan 16;385(2):393-404. doi: 10.1016/j.jmb.2008.11.003. Epub 2008 Nov 12. J Mol Biol. 2009. PMID: 19027026 Free PMC article.
-
Structural and functional characterisation of a conserved archaeal RadA paralog with antirecombinase activity.J Mol Biol. 2009 Jun 19;389(4):661-73. doi: 10.1016/j.jmb.2009.04.060. Epub 2009 May 3. J Mol Biol. 2009. PMID: 19414020 Free PMC article.
-
PDB_REDO: constructive validation, more than just looking for errors.Acta Crystallogr D Biol Crystallogr. 2012 Apr;68(Pt 4):484-96. doi: 10.1107/S0907444911054515. Epub 2012 Mar 16. Acta Crystallogr D Biol Crystallogr. 2012. PMID: 22505269 Free PMC article.
-
Detection of novel recombinases in bacteriophage genomes unveils Rad52, Rad51 and Gp2.5 remote homologs.Nucleic Acids Res. 2010 Jul;38(12):3952-62. doi: 10.1093/nar/gkq096. Epub 2010 Mar 1. Nucleic Acids Res. 2010. PMID: 20194117 Free PMC article.
References
-
- Grogan D.W. The question of DNA repair in hyperthermophilic archaea. Trends Microbiol. 2000;8:180–185. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

