NMR study of a membrane protein in detergent-free aqueous solution
- PMID: 15956183
- PMCID: PMC1157056
- DOI: 10.1073/pnas.0503750102
NMR study of a membrane protein in detergent-free aqueous solution
Abstract
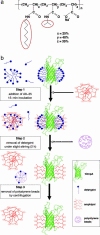
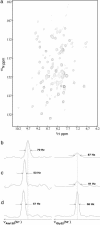
One of the major obstacles to membrane protein (MP) structural studies is the destabilizing effect of detergents. Amphipols (APols) are short amphipathic polymers that can substitute for detergents to keep MPs water-soluble under mild conditions. In the present work, we have explored the feasibility of studying the structure of APol-complexed MPs by NMR. As a test MP, we chose the 171-residue transmembrane domain of outer MP A from Escherichia coli (tOmpA), whose x-ray and NMR structures in detergent are known. 2H,15N-labeled tOmpA was produced as inclusion bodies, refolded in detergent solution, trapped with APol A8-35, and the detergent removed by adsorption onto polystyrene beads. The resolution of transverse relaxation-optimized spectroscopy-heteronuclear single-quantum correlation spectra of tOmpA/A8-35 complexes was found to be close to that of the best spectra obtained in detergent solutions. The dispersion of chemical shifts indicated that the protein had regained its native fold and retained it during the exchange of surfactants. MP-APol interactions were mapped by substituting hydrogenated for deuterated A8-35. The resulting dipolar broadening of amide proton linewidths was found to be limited to the beta-barrel region of tOmpA, indicating that A8-35 binds specifically to the hydrophobic transmembrane surface of the protein. The potential of this approach to MP studies by solution NMR is discussed.
Figures
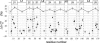

References
-
- le Maire, M., Champeil, P. & Møller, J. V. (2000) Biochim. Biophys. Acta 1508, 86–111. - PubMed
-
- Bowie, J. U. (2001) Curr. Opin. Struct. Biol. 11, 397–402. - PubMed
-
- McDonnell, P. A. & Opella, S. J. (1993) J. Magn. Reson. B102, 120–125.
-
- MacKenzie, K. R., Prestegard, J. H. & Engelman, D. M. (1997) Science 276, 131–133. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

