Use of the INNO-LiPA-MYCOBACTERIA assay (version 2) for identification of Mycobacterium avium-Mycobacterium intracellulare-Mycobacterium scrofulaceum complex isolates
- PMID: 15956365
- PMCID: PMC1151901
- DOI: 10.1128/JCM.43.6.2567-2574.2005
Use of the INNO-LiPA-MYCOBACTERIA assay (version 2) for identification of Mycobacterium avium-Mycobacterium intracellulare-Mycobacterium scrofulaceum complex isolates
Abstract
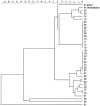
Using INNO-LiPA-MYCOBACTERIA (Lipav1; Innogenetics) and the AccuProbe (Gen-Probe Inc./bioMérieux) techniques, 35 Mycobacterium avium-Mycobacterium intracellulare-Mycobacterium scrofulaceum (MAC/MAIS) complex strains were identified between January 2000 and December 2002. Thirty-four of 35 isolates were positive only for the MAIS complex probe by Lipav1 and were further analyzed by INNO-LiPA-MYCOBACTERIA version 2 (Lipav2), hsp65 PCR restriction pattern analysis (PRA), and ribosomal internal transcribed spacer (ITS), hsp65, and 16S rRNA sequences. Lipav2 identified 14 of 34 strains at the species level, including 11 isolates positive for the newly specific MAC sequevar Mac-A probe (MIN-2 probe). Ten of these 11 isolates corresponded to sequevar Mac-A, which was recently defined as Mycobacterium chimerae sp. nov. Among the last 20 of the 34 MAIS isolates, 17 (by hsp65 PRA) and 18 (by hsp65 sequence) were characterized as M. avium. Ten of the 20 were identified as Mac-U sequevar. All these 20 isolates were identified as M. intracellulare by 16S rRNA sequence except one isolate identified as Mycobacterium paraffinicum by 16S rRNA and ITS sequencing. One isolate out of 35 isolates that was positive for M. avium by AccuProbe and that was Mycobacterium genus probe positive and MAIS probe negative by Lipav1 and Lipav2 might be considered a new species. In conclusion, the new INNO-LiPA-MYCOBACTERIA allowed the identification of 40% of the previously unidentified MAIS isolates at the species level. The results of the Lipav2 assay on the MAIS isolates confirm the great heterogeneity of this group and suggest the use of hsp65 or ITS sequencing for precise identification of such isolates.
Figures


References
-
- Alcaide, F., I. Richter, C. Bernasconi, B. Springer, C. Hagenau, R. Schulze-Röbbecke, E. Tortoli, R. Martin, E. C. Böttger, and A. Telenti. 1997. Heterogeneity and clonality among isolates of Mycobacterium kansasii: implications for epidemiological and pathogenicity studies. J. Clin. Microbiol. 35:1959-1964. - PMC - PubMed
-
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. - PubMed
-
- Böddinghaus, B., J. Wolters, W. Heikens, and E. C. Böttger. 1992. Phylogenetic analysis and identification of different serovars of Mycobacterium intracellulare at the molecular level. FEMS Microbiol. Lett. 70:197-204. - PubMed
-
- De Smet, K. A., T. J. Hellyer, A. W. Khan, I. N. Brown, and J. Ivanyi. 1996. Genetic and serovar typing of clinical isolates of the Mycobacterium avium-intracellulare complex. Tuberc. Lung Dis. 77:71-76. - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

