Molecular epidemiology of endemic Clostridium difficile infection and the significance of subtypes of the United Kingdom epidemic strain (PCR ribotype 1)
- PMID: 15956384
- PMCID: PMC1151908
- DOI: 10.1128/JCM.43.6.2685-2696.2005
Molecular epidemiology of endemic Clostridium difficile infection and the significance of subtypes of the United Kingdom epidemic strain (PCR ribotype 1)
Abstract
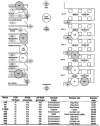
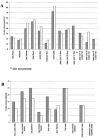
We previously identified two subtypes of the epidemic strain Clostridium difficile PCR ribotype 1, one clindamycin-sensitive strain (arbitrarily primed PCR [AP-PCR] type Ia) and a closely related clindamycin-resistant strain (AP-PCR type Ib) in our institution. We have now carried out prospective epidemiological surveillance for 4 years, immediately following the relocation of two acute medicine wards for elderly patients (wards A and B), to determine the clinical epidemiology of subtypes of the epidemic C. difficile PCR ribotype 1 group. To maximize the chance of strain discrimination, we used three DNA fingerprinting methods, AP-PCR, ribospacer PCR (RS-PCR), and pulsed-field gel electrophoresis (PFGE), to analyze C. difficile isolates recovered from symptomatic patients and from repeated environmental samplings. On ward B the incidence of C. difficile infection correlated significantly with the prevalence of environmental C. difficile both in ward areas closely associated with patients and health care personnel (r = 0.53; P < 0.05) and in high-reach sites (r = 0.85; P < 0.05). No such relationships were found on ward A. Seventeen distinct C. difficile genotypes were identified, 17 by AP-PCR, 12 by PFGE, and 11 by RS-PCR, but only 4 of 17 genotypes caused patient infection. Isolates recovered from the hospital ward environment were much more diverse (14 genotypes). AP-PCR type Ia represented >90% of the C. difficile isolates. In addition to this genotype, only two others were isolated from both patient feces and environmental surfaces. AP-PCR type Ib (clindamycin-resistant PCR ribotype 1 clone) was not associated with any cases of C. difficile infection and was isolated from the environment on only two occasions, after having been implicated in a cluster of six C. difficile infections 5 months before this study. The disappearance of this strain implies that differences in virulence and/or selective pressures may exist for this strain and the closely related, widespread C. difficile AP-PCR type Ia strain. Our findings emphasize the need to understand the epidemiology and virulence of clinically significant strains to determine successful control measures for C. difficile infections.
Figures




Similar articles
-
A modified pulsed-field gel electrophoresis (PFGE) protocol for subtyping previously non-PFGE typeable isolates of Clostridium difficile polymerase chain reaction ribotype 001.J Hosp Infect. 2005 Nov;61(3):231-6. doi: 10.1016/j.jhin.2005.01.017. Epub 2005 Jul 5. J Hosp Infect. 2005. PMID: 16002184
-
Analysis of Clostridium difficile isolates from nosocomial outbreaks at three hospitals in diverse areas of Japan.J Clin Microbiol. 2001 Apr;39(4):1391-5. doi: 10.1128/JCM.39.4.1391-1395.2001. J Clin Microbiol. 2001. PMID: 11283061 Free PMC article.
-
Molecular epidemiology of endemic Clostridium difficile infection.Epidemiol Infect. 2001 Jun;126(3):343-50. doi: 10.1017/s095026880100557x. Epidemiol Infect. 2001. PMID: 11467790 Free PMC article.
-
Typing of Clostridium difficile.Clin Microbiol Infect. 2001 Aug;7(8):428-31. doi: 10.1046/j.1198-743x.2001.00288.x. Clin Microbiol Infect. 2001. PMID: 11591206 Review.
-
Current application and future perspectives of molecular typing methods to study Clostridium difficile infections.Euro Surveill. 2013 Jan 24;18(4):20381. doi: 10.2807/ese.18.04.20381-en. Euro Surveill. 2013. PMID: 23369393 Review.
Cited by
-
Clostridium difficile PCR ribotype 027: assessing the risks of further worldwide spread.Lancet Infect Dis. 2010 Jun;10(6):395-404. doi: 10.1016/S1473-3099(10)70080-3. Lancet Infect Dis. 2010. PMID: 20510280 Free PMC article. Review.
-
Development and validation of an internationally-standardized, high-resolution capillary gel-based electrophoresis PCR-ribotyping protocol for Clostridium difficile.PLoS One. 2015 Feb 13;10(2):e0118150. doi: 10.1371/journal.pone.0118150. eCollection 2015. PLoS One. 2015. PMID: 25679978 Free PMC article.
-
Molecular analysis of Clostridium difficile at a university teaching hospital in Japan: a shift in the predominant type over a five-year period.Eur J Clin Microbiol Infect Dis. 2007 Oct;26(10):695-703. doi: 10.1007/s10096-007-0355-8. Eur J Clin Microbiol Infect Dis. 2007. PMID: 17647032
-
Clostridium difficile infection in non-HIV-immunocompromised patients and in HIV-infected patients.Curr Gastroenterol Rep. 2011 Aug;13(4):344-50. doi: 10.1007/s11894-011-0196-6. Curr Gastroenterol Rep. 2011. PMID: 21541693 Review.
-
Antimicrobial Resistance and Reduced Susceptibility in Clostridium difficile: Potential Consequences for Induction, Treatment, and Recurrence of C. difficile Infection.Antibiotics (Basel). 2015 Jul 10;4(3):267-98. doi: 10.3390/antibiotics4030267. Antibiotics (Basel). 2015. PMID: 27025625 Free PMC article. Review.
References
-
- Archibald, L. K., S. N. Banerjee, and W. R. Jarvis. 2004. Secular trends in hospital-acquired Clostridium difficile disease in the United States, 1987-2001. J. Infect. Dis. 189:1585-1589. - PubMed
-
- Barbut, F., G. Corthier, Y. Charpak, M. Cerf, H. Monteil, T. Fosse, A. Trevoux, B. De Barbeyrac, Y. Boussougant, S. Tigaud, F. Tytgat, A. Sedallian, S. Duborgel, A. Collignon, M. E. Le Guern, P. Bernasconi, and J. C. Petit. 1996. Prevalence and pathogenicity of Clostridium difficile in hospitalized patients. A French multicenter study. Arch. Intern. Med. 156:1449-1454. - PubMed
-
- Bartlett, J. G., T. W. Chang, M. J. Gurwith, S. L. Gorbach, and A. B. Onderdonk. 1978. Antibiotic associated pseudomembranous colitis due to toxin producing clostridia. N. Engl. J. Med. 298:531-534. - PubMed
-
- Brazier, J. S. 1998. The epidemiology and typing of Clostridium difficile. J. Antimicrob. Chemother. 41(Suppl. C):47-57. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

