Gene 5 of the avian coronavirus infectious bronchitis virus is not essential for replication
- PMID: 15956552
- PMCID: PMC1143771
- DOI: 10.1128/JVI.79.13.8065-8078.2005
Gene 5 of the avian coronavirus infectious bronchitis virus is not essential for replication
Abstract
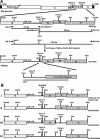
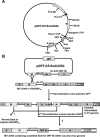
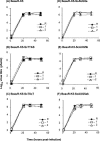
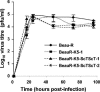
The avian coronavirus Infectious bronchitis virus (IBV), like other coronaviruses, expresses several small nonstructural (ns) proteins in addition to those from gene 1 (replicase) and the structural proteins. These coronavirus ns genes differ both in number and in amino acid similarity between the coronavirus groups but show some concordance within a group or subgroup. The functions and requirements of the small ns gene products remain to be elucidated. With the advent of reverse genetics for coronaviruses, the first steps in elucidating their role can be investigated. We have used our reverse genetics system for IBV (R. Casais, V. Thiel, S. G. Siddell, D. Cavanagh, and P. Britton, J. Virol. 75:12359-12369, 2001) to investigate the requirement of IBV gene 5 for replication in vivo, in ovo, and ex vivo. We produced a series of recombinant viruses, with an isogenic background, in which complete expression of gene 5 products was prevented by the inactivation of gene 5 following scrambling of the transcription-associated sequence, thereby preventing the expression of IBV subgenomic mRNA 5, or scrambling either separately or together of the translation initiation codons for the two gene 5 products. As all of the recombinant viruses replicated very similarly to the wild-type virus, Beau-R, we conclude that the IBV gene 5 products are not essential for IBV replication per se and that they are accessory proteins.
Figures

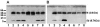


References
-
- Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1987. Current protocols in molecular biology. John Wiley and Sons, Inc., New York, N.Y.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

