Recombinant human immunodeficiency virus type 1 integrase exhibits a capacity for full-site integration in vitro that is comparable to that of purified preintegration complexes from virus-infected cells
- PMID: 15956566
- PMCID: PMC1143728
- DOI: 10.1128/JVI.79.13.8208-8216.2005
Recombinant human immunodeficiency virus type 1 integrase exhibits a capacity for full-site integration in vitro that is comparable to that of purified preintegration complexes from virus-infected cells
Abstract
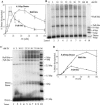
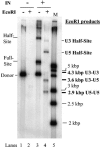
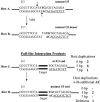
Retrovirus preintegration complexes (PIC) in virus-infected cells contain the linear viral DNA genome (approximately 10 kbp), viral proteins including integrase (IN), and cellular proteins. After transport of the PIC into the nucleus, IN catalyzes the concerted insertion of the two viral DNA ends into the host chromosome. This successful insertion process is termed "full-site integration." Reconstitution of nucleoprotein complexes using recombinant human immunodeficiency virus type 1 (HIV-1) IN and model viral DNA donor substrates (approximately 0.30 to 0.48 kbp in length) that are capable of catalyzing efficient full-site integration has proven difficult. Many of the products are half-site integration reactions where either IN inserts only one end of the viral donor substrate into a circular DNA target or into other donors. In this report, we have purified recombinant HIV-1 IN at pH 6.8 in the presence of MgSO4 that performed full-site integration nearly as efficiently as HIV-1 PIC. The size of the viral DNA substrate was significantly increased to 4.1 kbp, thus allowing for the number of viral DNA ends and the concentrations of IN in the reaction mixtures to be decreased by a factor of approximately 10. In a typical reaction at 37 degrees C, recombinant HIV-1 IN at 5 to 10 nM incorporated 30 to 40% of the input DNA donor into full-site integration products. The synthesis of full-site products continued up to approximately 2 h, comparable to incubation times used with HIV-1 PIC. Approximately 5% of the input donor was incorporated into the circular target producing half-site products with no significant quantities of other integration products produced. DNA sequence analysis of the viral DNA-target junctions derived from wild-type U3 and U5 coupled reactions showed an approximately 70% fidelity for the HIV-1 5-bp host site duplications. Recombinant HIV-1 IN successfully utilized a mutant U5 end containing additional nucleotide extensions for full-site integration demonstrating that IN worked properly under nonideal active substrate conditions. The fidelity of the 5-bp host site duplications was also high with these coupled mutant U5 and wild-type U3 donor ends. These studies suggest that recombinant HIV-1 IN is at least as capable as native IN in virus particles and approaching that observed with HIV-1 PIC for catalyzing full-site integration.
Figures




References
-
- Anthony, N. J. 2004. HIV-1 integrase: a target for new AIDS chemotherapeutics. Curr. Top. Med. Chem. 4:979-990. - PubMed
-
- Brin, E., and J. Leis. 2002. Changes in the mechanism of DNA integration in vitro induced by base substitutions in the HIV-1 U5 and U3 terminal sequences. J. Biol. Chem. 277:10938-10948. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

