New DNA viruses identified in patients with acute viral infection syndrome
- PMID: 15956568
- PMCID: PMC1143717
- DOI: 10.1128/JVI.79.13.8230-8236.2005
New DNA viruses identified in patients with acute viral infection syndrome
Abstract
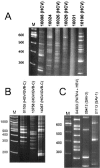
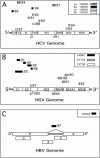
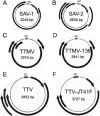
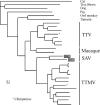
A sequence-independent PCR amplification method was used to identify viral nucleic acids in the plasma samples of 25 individuals presenting with symptoms of acute viral infection following high-risk behavior for human immunodeficiency virus type 1 transmission. GB virus C/hepatitis G virus was identified in three individuals and hepatitis B virus in one individual. Three previously undescribed DNA viruses were also detected, a parvovirus and two viruses related to TT virus (TTV). Nucleic acids in human plasma that were distantly related to bacterial sequences or with no detectable similarities to known sequences were also found. Nearly complete viral genome sequencing and phylogenetic analysis confirmed the presence of a new parvovirus distinct from known human and animal parvoviruses and of two related TTV-like viruses highly divergent from both the TTV and TTV-like minivirus groups. The detection of two previously undescribed viral species in a small group of individuals presenting acute viral syndrome with unknown etiology indicates that a rich yield of new human viruses may be readily identifiable using simple methods of sequence-independent nucleic acid amplification and limited sequencing.
Figures


References
-
- Baylis, S. A., N. Shah, and P. D. Minor. 2004. Evaluation of different assays for the detection of parvovirus B19 DNA in human plasma. J. Virol. Methods 121:7-16. - PubMed
-
- Burnouf, T., and M. Radosevich. 2000. Reducing the risk of infection from plasma products: specific preventative strategies. Blood Rev. 14:94-110. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

