Role of N-linked glycans in the functions of hepatitis C virus envelope glycoproteins
- PMID: 15956584
- PMCID: PMC1143753
- DOI: 10.1128/JVI.79.13.8400-8409.2005
Role of N-linked glycans in the functions of hepatitis C virus envelope glycoproteins
Abstract
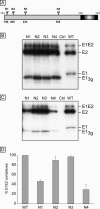
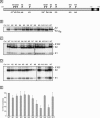
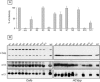
Hepatitis C virus (HCV) encodes two viral envelope glycoproteins. E1 contains 4 or 5 N-linked glycosylation sites and E2 contains up to 11, with most of the sites being well conserved, suggesting that they play an essential role in some functions of these proteins. For this study, we used retroviral pseudotyped particles harboring mutated HCV envelope glycoproteins to study these glycans. The mutants were named with an N followed by a number related to the relative position of the potential glycosylation site in each glycoprotein (E1N1 to E1N4 for E1 mutants and E2N1 to E2N11 for E2 mutants). The characterization of these mutants allowed us to define three phenotypes. For the first group (E1N3, E2N3, E2N5, E2N6, E2N7, and E2N9), the infectivities of the mutants were close to that of the wild type. The second group (E1N1, E1N2, E1N4, E2N1, and E2N11) contained mutants that were still infectious but whose infectivities were reduced to <50% that of the wild type. The third group (E2N2, E2N4, E2N8, and E2N10) contained mutants that had almost totally lost infectivity. The absence of infectivity of the E2N8 and E2N10 mutants was due to the lack of incorporation of the E1E2 heterodimer into HCVpp, which was due to misfolding of the heterodimer, as shown by immunoprecipitation with conformation-sensitive antibodies and by a CD81 pull-down assay. The absence of infectivity of the E2N2 and E2N4 mutants indicated that these two glycans are involved in controlling HCV entry. Altogether, the data indicate that some glycans of HCV envelope glycoproteins play a major role in protein folding and others play a role in HCV entry.
Figures


References
-
- Ausubel, F. M., R. Brent, R. E. Kingston, A. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 2000. Current protocols in molecular biology, vol. 1. John Wiley and Sons, Inc., New York, N.Y.
-
- Bashirova, A. A., T. B. Geijtenbeek, G. C. van Duijnhoven, S. J. van Vliet, J. B. Eilering, M. P. Martin, L. Wu, T. D. Martin, N. Viebig, P. A. Knolle, V. N. KewalRamani, Y. van Kooyk, and M. Carrington. 2001. A dendritic cell-specific intercellular adhesion molecule 3-grabbing nonintegrin (DC-SIGN)-related protein is highly expressed on human liver sinusoidal endothelial cells and promotes HIV-1 infection. J. Exp. Med. 193:671-678. - PMC - PubMed
-
- Bause, E., and H. Hettkamp. 1979. Primary structural requirements for N-glycosylation of peptides in rat liver. FEBS Lett. 108:341-344. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

