Genetic regulation of gene expression during shoot development in Arabidopsis
- PMID: 15956669
- PMCID: PMC1456214
- DOI: 10.1534/genetics.105.042275
Genetic regulation of gene expression during shoot development in Arabidopsis
Abstract
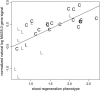
The genetic control of gene expression during shoot development in Arabidopsis thaliana was analyzed by combining quantitative trait loci (QTL) and microarray analysis. Using oligonucleotide array data from 30 recombinant inbred lines derived from a cross of Columbia and Landsberg erecta ecotypes, the Arabidopsis genome was scanned for marker-by-gene linkages or so-called expression QTL (eQTL). Single-feature polymorphisms (SFPs) associated with sequence disparities between ecotypes were purged from the data. SFPs may alter the hybridization efficiency between cDNAs from one ecotype with probes of another ecotype. In genome scans, five eQTL hot spots were found with significant marker-by-gene linkages. Two of the hot spots coincided with classical QTL conditioning shoot regeneration, suggesting that some of the heritable gene expression changes observed in this study are related to differences in shoot regeneration efficiency between ecotypes. Some of the most significant eQTL, particularly those at the shoot regeneration QTL sites, tended to show cis-chromosomal linkages in that the target genes were located at or near markers to which their expression was linked. However, many linkages of lesser significance showed expected "trans-effects," whereby a marker affects the expression of a target gene located elsewhere on the genome. Some of these eQTL were significantly linked to numerous genes throughout the genome, suggesting the occurrence of large groups of coregulated genes controlled by single markers.
Figures





References
-
- Affymetrix, 2002. Affymetrix Microarray Suite User Guide. Affymetrix, Santa Clara, CA.
-
- Anscombe, F. J., 1948. The transformation of Poisson, binomial and negative-binomial data. Biometrika 35: 246–254.
-
- Baurle, I., and T. Laux, 2003. Apical meristems: the plant's fountain of youth. BioEssays 25: 961–970. - PubMed
-
- Brem, R. B., G. Yvert, R. Clinton and L. Kruglyak, 2002. Genetic dissection of transcriptional regulation in budding yeast. Science 296: 752–755. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

