Comparative 3-D modeling of tmRNA
- PMID: 15958166
- PMCID: PMC1168896
- DOI: 10.1186/1471-2199-6-14
Comparative 3-D modeling of tmRNA
Abstract
Background: Trans-translation releases stalled ribosomes from truncated mRNAs and tags defective proteins for proteolytic degradation using transfer-messenger RNA (tmRNA). This small stable RNA represents a hybrid of tRNA- and mRNA-like domains connected by a variable number of pseudoknots. Comparative sequence analysis of tmRNAs found in bacteria, plastids, and mitochondria provides considerable insights into their secondary structures. Progress toward understanding the molecular mechanism of template switching, which constitutes an essential step in trans-translation, is hampered by our limited knowledge about the three-dimensional folding of tmRNA.
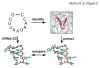
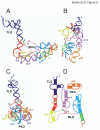
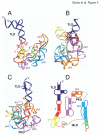
Results: To facilitate experimental testing of the molecular intricacies of trans-translation, which often require appropriately modified tmRNA derivatives, we developed a procedure for building three-dimensional models of tmRNA. Using comparative sequence analysis, phylogenetically-supported 2-D structures were obtained to serve as input for the program ERNA-3D. Motifs containing loops and turns were extracted from the known structures of other RNAs and used to improve the tmRNA models. Biologically feasible 3-D models for the entire tmRNA molecule could be obtained. The models were characterized by a functionally significant close proximity between the tRNA-like domain and the resume codon. Potential conformational changes which might lead to a more open structure of tmRNA upon binding to the ribosome are discussed. The method, described in detail for the tmRNAs of Escherichia coli, Bacillus anthracis, and Caulobacter crescentus, is applicable to every tmRNA.
Conclusion: Improved molecular models of biological significance were obtained. These models will guide in the design of experiments and provide a better understanding of trans-translation. The comparative procedure described here for tmRNA is easily adopted for the modeling the members of other RNA families.
Figures



Similar articles
-
Requirements for resuming translation in chimeric transfer-messenger RNAs of Escherichia coli and Mycobacterium tuberculosis.BMC Mol Biol. 2014 Sep 15;15:19. doi: 10.1186/1471-2199-15-19. BMC Mol Biol. 2014. PMID: 25220282 Free PMC article.
-
Escherichia coli tmRNA lacking pseudoknot 1 tags truncated proteins in vivo and in vitro.RNA. 2009 Jan;15(1):128-37. doi: 10.1261/rna.1192409. Epub 2008 Nov 10. RNA. 2009. PMID: 19001120 Free PMC article.
-
Transfer-messenger RNA unfolds as it transits the ribosome.RNA. 2005 May;11(5):668-73. doi: 10.1261/rna.7269305. Epub 2005 Apr 5. RNA. 2005. PMID: 15811920 Free PMC article.
-
A salvage pathway for protein structures: tmRNA and trans-translation.Annu Rev Microbiol. 2003;57:101-23. doi: 10.1146/annurev.micro.57.030502.090945. Epub 2003 May 1. Annu Rev Microbiol. 2003. PMID: 12730326 Review.
-
Quality control of the elongation step of protein synthesis by tmRNP.J Nutr. 2001 Nov;131(11):2978S-82S. doi: 10.1093/jn/131.11.2978S. J Nutr. 2001. PMID: 11694632 Review.
Cited by
-
A Proposal for the RNAome at the Dawn of the Last Universal Common Ancestor.Genes (Basel). 2024 Sep 11;15(9):1195. doi: 10.3390/genes15091195. Genes (Basel). 2024. PMID: 39336786 Free PMC article.
-
The role of upstream sequences in selecting the reading frame on tmRNA.BMC Biol. 2008 Jun 30;6:29. doi: 10.1186/1741-7007-6-29. BMC Biol. 2008. PMID: 18590561 Free PMC article.
-
In vivo tmRNA protection by SmpB and pre-ribosome binding conformation in solution.RNA. 2014 Oct;20(10):1607-20. doi: 10.1261/rna.045674.114. Epub 2014 Aug 18. RNA. 2014. PMID: 25135523 Free PMC article.
-
Detection of tmRNA molecules on microarrays at low temperatures using helper oligonucleotides.BMC Biotechnol. 2010 Apr 28;10:34. doi: 10.1186/1472-6750-10-34. BMC Biotechnol. 2010. PMID: 20426847 Free PMC article.
-
The tmRDB and SRPDB resources.Nucleic Acids Res. 2006 Jan 1;34(Database issue):D163-8. doi: 10.1093/nar/gkj142. Nucleic Acids Res. 2006. PMID: 16381838 Free PMC article.
References
-
- Keiler KC, Waller PR, Sauer RT. Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA. Science. 1996;271:990–993. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

