DNA knots reveal a chiral organization of DNA in phage capsids
- PMID: 15958528
- PMCID: PMC1166588
- DOI: 10.1073/pnas.0409323102
DNA knots reveal a chiral organization of DNA in phage capsids
Abstract
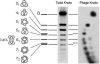
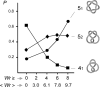
Icosahedral bacteriophages pack their double-stranded DNA genomes to near-crystalline density and achieve one of the highest levels of DNA condensation found in nature. Despite numerous studies, some essential properties of the packaging geometry of the DNA inside the phage capsid are still unknown. We present a different approach to the problems of randomness and chirality of the packed DNA. We recently showed that most DNA molecules extracted from bacteriophage P4 are highly knotted because of the cyclization of the linear DNA molecule confined in the phage capsid. Here, we show that these knots provide information about the global arrangement of the DNA inside the capsid. First, we analyze the distribution of the viral DNA knots by high-resolution gel electrophoresis. Next, we perform Monte Carlo computer simulations of random knotting for freely jointed polygons confined to spherical volumes. Comparison of the knot distributions obtained by both techniques produces a topological proof of nonrandom packaging of the viral DNA. Moreover, our simulations show that the scarcity of the achiral knot 4(1) and the predominance of the torus knot 5(1) over the twist knot 5(2) observed in the viral distribution of DNA knots cannot be obtained by confinement alone but must include writhe bias in the conformation sampling. These results indicate that the packaging geometry of the DNA inside the viral capsid is writhe-directed.
Figures


References
-
- Earnshaw, W. C. & Casjens, S. R. (1980) Cell 21, 319-331. - PubMed
-
- Rishov, S., Holzenburg, A., Johansen, B. V. & Lindqvist, B. H. (1998) Virology 245, 11-17. - PubMed
-
- Smith, D. E., Tans, S. J., Smith, S. B., Grimes S., Anderson, D. L. & Bustamante, C. (2001) Nature 413, 748-752. - PubMed
-
- Kellenberger, E., Carlemalm, E., Sechaud, J., Ryter, A. & Haller, G. (1986) in Bacterial Chromatin, eds. Gualerzi, C. & Pon, C. L. (Springer, Berlin), p. 11.
-
- San Martin, C. & Burnett, R. (2003) Curr. Top. Microbiol. Immunol. 272, 57-94. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

