Large-scale 13C-flux analysis reveals mechanistic principles of metabolic network robustness to null mutations in yeast
- PMID: 15960801
- PMCID: PMC1175969
- DOI: 10.1186/gb-2005-6-6-r49
Large-scale 13C-flux analysis reveals mechanistic principles of metabolic network robustness to null mutations in yeast
Abstract
Background: Quantification of intracellular metabolite fluxes by 13C-tracer experiments is maturing into a routine higher-throughput analysis. The question now arises as to which mutants should be analyzed. Here we identify key experiments in a systems biology approach with a genome-scale model of Saccharomyces cerevisiae metabolism, thereby reducing the workload for experimental network analyses and functional genomics.
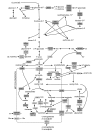
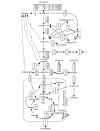
Results: Genome-scale 13C flux analysis revealed that about half of the 745 biochemical reactions were active during growth on glucose, but that alternative pathways exist for only 51 gene-encoded reactions with significant flux. These flexible reactions identified in silico are key targets for experimental flux analysis, and we present the first large-scale metabolic flux data for yeast, covering half of these mutants during growth on glucose. The metabolic lesions were often counteracted by flux rerouting, but knockout of cofactor-dependent reactions, as in the adh1, ald6, cox5A, fum1, mdh1, pda1, and zwf1 mutations, caused flux responses in more distant parts of the network. By integrating computational analyses, flux data, and physiological phenotypes of all mutants in active reactions, we quantified the relative importance of 'genetic buffering' through alternative pathways and network redundancy through duplicate genes for genetic robustness of the network.
Conclusions: The apparent dispensability of knockout mutants with metabolic function is explained by gene inactivity under a particular condition in about half of the cases. For the remaining 207 viable mutants of active reactions, network redundancy through duplicate genes was the major (75%) and alternative pathways the minor (25%) molecular mechanism of genetic network robustness in S. cerevisiae.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

