Refinement and prediction of protein prenylation motifs
- PMID: 15960807
- PMCID: PMC1175975
- DOI: 10.1186/gb-2005-6-6-r55
Refinement and prediction of protein prenylation motifs
Abstract
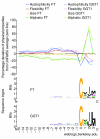
We refined the motifs for carboxy-terminal protein prenylation by analysis of known substrates for farnesyltransferase (FT), geranylgeranyltransferase I (GGT1) and geranylgeranyltransferase II (GGT2). In addition to the CaaX box for the first two enzymes, we identify a preceding linker region that appears constrained in physicochemical properties, requiring small or flexible, preferably hydrophilic, amino acids. Predictors were constructed on the basis of sequence and physical property profiles, including interpositional correlations, and are available as the Prenylation Prediction Suite (PrePS, http://mendel.imp.univie.ac.at/sat/PrePS) which also allows evaluation of evolutionary motif conservation. PrePS can predict partially overlapping substrate specificities, which is of medical importance in the case of understanding cellular action of FT inhibitors as anticancer and anti-parasite agents.
Figures
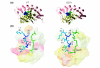



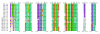
Similar articles
-
Towards complete sets of farnesylated and geranylgeranylated proteins.PLoS Comput Biol. 2007 Apr 6;3(4):e66. doi: 10.1371/journal.pcbi.0030066. Epub 2007 Feb 23. PLoS Comput Biol. 2007. PMID: 17411337 Free PMC article.
-
Protein prenyltransferases.Genome Biol. 2003;4(4):212. doi: 10.1186/gb-2003-4-4-212. Epub 2003 Apr 1. Genome Biol. 2003. PMID: 12702202 Free PMC article. Review.
-
N-terminal N-myristoylation of proteins: refinement of the sequence motif and its taxon-specific differences.J Mol Biol. 2002 Apr 5;317(4):523-40. doi: 10.1006/jmbi.2002.5425. J Mol Biol. 2002. PMID: 11955007
-
Crystallographic analysis of CaaX prenyltransferases complexed with substrates defines rules of protein substrate selectivity.J Mol Biol. 2004 Oct 15;343(2):417-33. doi: 10.1016/j.jmb.2004.08.056. J Mol Biol. 2004. PMID: 15451670
-
Advances in the development of farnesyltransferase inhibitors: substrate recognition by protein farnesyltransferase.J Cell Biochem Suppl. 1997;27:12-9. J Cell Biochem Suppl. 1997. PMID: 9591188 Review.
Cited by
-
Expansion of protein farnesyltransferase specificity using "tunable" active site interactions: development of bioengineered prenylation pathways.J Biol Chem. 2012 Nov 2;287(45):38090-100. doi: 10.1074/jbc.M112.404954. Epub 2012 Sep 19. J Biol Chem. 2012. PMID: 22992747 Free PMC article.
-
Brucella effectors NyxA and NyxB target SENP3 to modulate the subcellular localisation of nucleolar proteins.Nat Commun. 2023 Jan 6;14(1):102. doi: 10.1038/s41467-022-35763-8. Nat Commun. 2023. PMID: 36609656 Free PMC article.
-
ANNIE: integrated de novo protein sequence annotation.Nucleic Acids Res. 2009 Jul;37(Web Server issue):W435-40. doi: 10.1093/nar/gkp254. Epub 2009 Apr 23. Nucleic Acids Res. 2009. PMID: 19389726 Free PMC article.
-
Molecular characterization and identification of target protein of an important vesicle trafficking gene AlRab7 from a salt excreting halophyte Aeluropus lagopoides.DNA Cell Biol. 2015 Feb;34(2):83-91. doi: 10.1089/dna.2014.2592. Epub 2014 Nov 19. DNA Cell Biol. 2015. PMID: 25408252 Free PMC article.
-
Identification of novel peptide substrates for protein farnesyltransferase reveals two substrate classes with distinct sequence selectivities.J Mol Biol. 2010 Jan 8;395(1):176-90. doi: 10.1016/j.jmb.2009.10.038. Epub 2009 Oct 28. J Mol Biol. 2010. PMID: 19878682 Free PMC article.
References
-
- Caplin BE, Hettich LA, Marshall MS. Substrate characterization of the Saccharomyces cerevisiae protein farnesyltransferase and type-I protein geranylgeranyltransferase. Biochim Biophys Acta. 1994;1205:39–48. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

