Monitoring tat peptide binding to TAR RNA by solid-state 31P-19F REDOR NMR
- PMID: 15961729
- PMCID: PMC1151589
- DOI: 10.1093/nar/gki626
Monitoring tat peptide binding to TAR RNA by solid-state 31P-19F REDOR NMR
Abstract
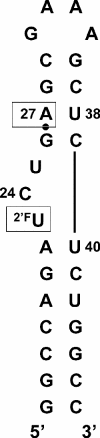
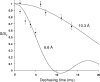
Complexes of the HIV transactivation response element (TAR) RNA with the viral regulatory protein tat are of special interest due in particular to the plasticity of the RNA at this binding site and to the potential for therapeutic targeting of the interaction. We performed REDOR solid-state NMR experiments on lyophilized samples of a 29 nt HIV-1 TAR construct to measure conformational changes in the tat-binding site concomitant with binding of a short peptide comprising the residues of the tat basic binding domain. Peptide binding was observed to produce a nearly 4 A decrease in the separation between phosphorothioate and 2'F labels incorporated at A27 in the upper helix and U23 in the bulge, respectively, consistent with distance changes observed in previous solution NMR studies, and with models showing significant rearrangement in position of bulge residue U23 in the bound-form RNA. In addition to providing long-range constraints on free TAR and the TAR-tat complex, these results suggest that in RNAs known to undergo large deformations upon ligand binding, 31P-19F REDOR measurements can also serve as an assay for complex formation in solid-state samples. To our knowledge, these experiments provide the first example of a solid-state NMR distance measurement in an RNA-peptide complex.
Figures



Similar articles
-
High affinity binding of TAR RNA by the human immunodeficiency virus type-1 tat protein requires base-pairs in the RNA stem and amino acid residues flanking the basic region.J Mol Biol. 1993 Mar 5;230(1):90-110. doi: 10.1006/jmbi.1993.1128. J Mol Biol. 1993. PMID: 8450553
-
Characterization of the solution conformations of unbound and Tat peptide-bound forms of HIV-1 TAR RNA.Biochemistry. 1999 Aug 3;38(31):10059-69. doi: 10.1021/bi990590h. Biochemistry. 1999. PMID: 10433713
-
13C/15N-19F intermolecular REDOR NMR study of the interaction of TAR RNA with Tat peptides.J Am Chem Soc. 2010 Dec 22;132(50):17643-5. doi: 10.1021/ja1051439. Epub 2010 Nov 24. J Am Chem Soc. 2010. PMID: 21105680 Free PMC article.
-
Discoveries of Tat-TAR interaction inhibitors for HIV-1.Curr Drug Targets Infect Disord. 2005 Dec;5(4):433-44. doi: 10.2174/156800505774912901. Curr Drug Targets Infect Disord. 2005. PMID: 16535863 Review.
-
Biochemical and functional interactions between HIV-1 Tat protein and TAR RNA.Arch Biochem Biophys. 1999 May 15;365(2):175-85. doi: 10.1006/abbi.1999.1206. Arch Biochem Biophys. 1999. PMID: 10328810 Review.
Cited by
-
Magic angle spinning NMR of viruses.Prog Nucl Magn Reson Spectrosc. 2015 Apr;86-87:21-40. doi: 10.1016/j.pnmrs.2015.02.003. Epub 2015 Feb 16. Prog Nucl Magn Reson Spectrosc. 2015. PMID: 25919197 Free PMC article. Review.
-
5-Fluoro pyrimidines: labels to probe DNA and RNA secondary structures by 1D 19F NMR spectroscopy.Nucleic Acids Res. 2009 Dec;37(22):7728-40. doi: 10.1093/nar/gkp862. Nucleic Acids Res. 2009. PMID: 19843610 Free PMC article.
-
Studying DNA G-Quadruplex Aptamer by 19F NMR.ACS Omega. 2017 Dec 31;2(12):8843-8848. doi: 10.1021/acsomega.7b01405. Epub 2017 Dec 12. ACS Omega. 2017. PMID: 30023592 Free PMC article.
-
Advances in solid-state NMR methods for studying RNA structures and dynamics.Magn Reson Lett. 2024 Apr 26;5(1):200133. doi: 10.1016/j.mrl.2024.200133. eCollection 2025 Feb. Magn Reson Lett. 2024. PMID: 40918042 Free PMC article. Review.
-
New, extended hairpin form of the TAR-2 RNA domain points to the structural polymorphism at the 5' end of the HIV-2 leader RNA.Nucleic Acids Res. 2006 May 31;34(10):2984-97. doi: 10.1093/nar/gkl373. Print 2006. Nucleic Acids Res. 2006. PMID: 16738137 Free PMC article.
References
-
- Leulliot N., Varani G. Current topics in RNA–protein recognition: control of specificity and biological function through induced fit and conformational capture. Biochemistry. 2001;40:7947–7956. - PubMed
-
- Puglisi J.D., Tan R., Calnan B.J., Frankel A.D., Williamson J.R. Conformation of the TAR RNA–arginine complex by NMR spectroscopy. Science. 1992;257:76–80. - PubMed
-
- Aboul-ela F., Karn J., Varani G. The structure of the human immunodeficiency virus type-1 TAR RNA reveals principles of RNA recognition by Tat protein. J. Mol. Biol. 1995;253:313–332. - PubMed
-
- Brodsky A.S., Williamson J.R. Solution structure of the HIV-2 TAR-argininamide complex. J. Mol. Biol. 1997;267:624–639. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

