Sprouty2 acts at the Cbl/CIN85 interface to inhibit epidermal growth factor receptor downregulation
- PMID: 15962011
- PMCID: PMC1369112
- DOI: 10.1038/sj.embor.7400453
Sprouty2 acts at the Cbl/CIN85 interface to inhibit epidermal growth factor receptor downregulation
Abstract
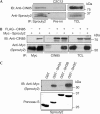
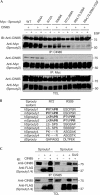
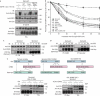
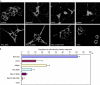
The ubiquitin ligase Cbl mediates ubiquitination of activated receptor tyrosine kinases (RTKs) and interacts with endocytic scaffold complexes, including CIN85/endophilins, to facilitate RTK endocytosis and degradation. Several mechanisms regulate the functions of Cbl to ensure the fine-tuning of RTK signalling and cellular homeostasis. One regulatory mechanism involves the binding of Cbl to Sprouty2, which sequesters Cbl away from activated epidermal growth factor receptors (EGFRs). Here, we show that Sprouty2 associates with CIN85 and acts at the interface between Cbl and CIN85 to inhibit EGFR downregulation. The CIN85 SH3 domains A and C bind specifically to proline-arginine motifs present in Sprouty2. Intact association between Sprouty2, Cbl and CIN85 is required for inhibition of EGFR endocytosis as well as EGF-induced differentiation of PC12 cells. Moreover, Sprouty4, which lacks CIN85-binding sites, does not inhibit EGFR downregulation, providing a molecular explanation for functional differences between Sprouty isoforms. Sprouty2 therefore acts as an inducible inhibitor of EGFR downregulation by targeting both the Cbl and CIN85 pathways.
Figures

Similar articles
-
Identification of a novel proline-arginine motif involved in CIN85-dependent clustering of Cbl and down-regulation of epidermal growth factor receptors.J Biol Chem. 2003 Oct 10;278(41):39735-46. doi: 10.1074/jbc.M304541200. Epub 2003 Jul 21. J Biol Chem. 2003. PMID: 12874286
-
Alix/AIP1 antagonizes epidermal growth factor receptor downregulation by the Cbl-SETA/CIN85 complex.Mol Cell Biol. 2004 Oct;24(20):8981-93. doi: 10.1128/MCB.24.20.8981-8993.2004. Mol Cell Biol. 2004. PMID: 15456872 Free PMC article.
-
Tyrosine phosphorylation of Sprouty proteins regulates their ability to inhibit growth factor signaling: a dual feedback loop.Mol Biol Cell. 2004 May;15(5):2176-88. doi: 10.1091/mbc.e03-07-0503. Epub 2004 Mar 5. Mol Biol Cell. 2004. Retraction in: Mol Biol Cell. 2022 Jul 1;33(8):re3. doi: 10.1091/mbc.E03-07-0503-corr. PMID: 15004239 Free PMC article. Retracted.
-
[Ubiquitination-mediated degradation of epidermal growth factor receptor].Zhongguo Yi Xue Ke Xue Yuan Xue Bao. 2005 Feb;27(1):120-7. Zhongguo Yi Xue Ke Xue Yuan Xue Bao. 2005. PMID: 15782507 Review. Chinese.
-
Negative regulation of receptor tyrosine kinases by ubiquitination: Key roles of the Cbl family of E3 ubiquitin ligases.Front Endocrinol (Lausanne). 2022 Jul 28;13:971162. doi: 10.3389/fendo.2022.971162. eCollection 2022. Front Endocrinol (Lausanne). 2022. PMID: 35966060 Free PMC article. Review.
Cited by
-
A novel role of Sprouty 2 in regulating cellular apoptosis.J Biol Chem. 2008 Feb 8;283(6):3181-3190. doi: 10.1074/jbc.M706567200. Epub 2007 Dec 10. J Biol Chem. 2008. PMID: 18070883 Free PMC article.
-
Regulation of cellular levels of Sprouty2 protein by prolyl hydroxylase domain and von Hippel-Lindau proteins.J Biol Chem. 2011 Dec 9;286(49):42027-42036. doi: 10.1074/jbc.M111.303222. Epub 2011 Oct 17. J Biol Chem. 2011. PMID: 22006925 Free PMC article.
-
Sprouty1 is a broad mediator of cellular senescence.Cell Death Dis. 2024 Apr 26;15(4):296. doi: 10.1038/s41419-024-06689-4. Cell Death Dis. 2024. PMID: 38670941 Free PMC article.
-
Threshold-controlled ubiquitination of the EGFR directs receptor fate.EMBO J. 2013 Jul 31;32(15):2140-57. doi: 10.1038/emboj.2013.149. Epub 2013 Jun 25. EMBO J. 2013. PMID: 23799367 Free PMC article.
-
Sprouty 2 regulates DNA damage-induced apoptosis in Ras-transformed human fibroblasts.J Biol Chem. 2009 Jan 9;284(2):848-54. doi: 10.1074/jbc.M808045200. Epub 2008 Nov 13. J Biol Chem. 2009. PMID: 19008219 Free PMC article.
References
-
- Christofori G (2003) Split personalities: the agonistic antagonist Sprouty. Nat Cell Biol 5: 377–379 - PubMed
-
- Dikic I, Giordano S (2003) Negative receptor signalling. Curr Opin Cell Biol 15: 128–135 - PubMed
-
- Fong CW, Leong HF, Wong ES, Lim J, Yusoff P, Guy GR (2003) Tyrosine phosphorylation of Sprouty2 enhances its interaction with c-Cbl and is crucial for its function. J Biol Chem 278: 33456–33464 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

