Deletions involving long-range conserved nongenic sequences upstream and downstream of FOXL2 as a novel disease-causing mechanism in blepharophimosis syndrome
- PMID: 15962237
- PMCID: PMC1224524
- DOI: 10.1086/432083
Deletions involving long-range conserved nongenic sequences upstream and downstream of FOXL2 as a novel disease-causing mechanism in blepharophimosis syndrome
Abstract
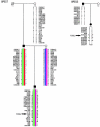
The expression of a gene requires not only a normal coding sequence but also intact regulatory regions, which can be located at large distances from the target genes, as demonstrated for an increasing number of developmental genes. In previous mutation studies of the role of FOXL2 in blepharophimosis syndrome (BPES), we identified intragenic mutations in 70% of our patients. Three translocation breakpoints upstream of FOXL2 in patients with BPES suggested a position effect. Here, we identified novel microdeletions outside of FOXL2 in cases of sporadic and familial BPES. Specifically, four rearrangements, with an overlap of 126 kb, are located 230 kb upstream of FOXL2, telomeric to the reported translocation breakpoints. Moreover, the shortest region of deletion overlap (SRO) contains several conserved nongenic sequences (CNGs) harboring putative transcription-factor binding sites and representing potential long-range cis-regulatory elements. Interestingly, the human region orthologous to the 12-kb sequence deleted in the polled intersex syndrome in goat, which is an animal model for BPES, is contained in this SRO, providing evidence of human-goat conservation of FOXL2 expression and of the mutational mechanism. Surprisingly, in a fifth family with BPES, one rearrangement was found downstream of FOXL2. In addition, we report nine novel rearrangements encompassing FOXL2 that range from partial gene deletions to submicroscopic deletions. Overall, genomic rearrangements encompassing or outside of FOXL2 account for 16% of all molecular defects found in our families with BPES. In summary, this is the first report of extragenic deletions in BPES, providing further evidence of potential long-range cis-regulatory elements regulating FOXL2 expression. It contributes to the enlarging group of developmental diseases caused by defective distant regulation of gene expression. Finally, we demonstrate that CNGs are candidate regions for genomic rearrangements in developmental genes.
Figures



References
Web Resources
-
- BACPAC Resources Center, http://bacpac.chori.org/
-
- Biomax Human Genome Database, http://www.biomax.de/products/bhgdb/bhgdb.htm
-
- ClustalW, http://www.ebi.ac.uk/clustalw/
-
- Find Simple Repeats software, http://zeon.well.ox.ac.uk/git-bin/microsatellite.cgi
-
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for PIS locus [accession number AF404302] and 48-kb goat sequence [accession number AF404302])
References
-
- Balemans W, Patel N, Ebeling M, Van Hul E, Wuyts W, Lacza C, Dioszegi M, Dikkers FG, Hildering P, Willems PJ, Verheij JB, Lindpaintner K, Vickery B, Foernzler D, Van Hul W (2002) Identification of a 52 kb deletion downstream of the SOST gene in patients with van Buchem disease. J Med Genet 39:91–97 - PMC - PubMed
-
- Bejerano G, Pheasant M, Makunin I, Stephen S, Kent WJ, Mattick JS, Haussler D (2004) Ultraconserved elements in the human genome. Science 304:1321–1325 - PubMed
-
- Bodega B, Porta C, Crosignani PG, Ginelli E, Marozzi A (2004) Mutations in the coding region of the FOXL2 gene are not a major cause of idiopathic premature ovarian failure. Mol Hum Reprod 10:555–557 - PubMed
-
- Cha SC, Jang YS, Lee JH, Kim HK, Kim SC, Kim S, Baek SH, Jung WS, Kim JR (2003) Mutational analysis of forkhead transcriptional factor 2 (FOXL2) in Korean patients with blepharophimosis-ptosis-epicanthus inversus syndrome. Clin Genet 64:485–490 - PubMed
-
- Cocquet J, De Baere E, Gareil M, Pannetier M, Xia X, Fellous M, Veitia RA (2003) Structure, evolution and expression of the FOXL2 transcription unit. Cytogenet Genome Res 101:206–211 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

