Cognate peptide-receptor ligand mapping by directed phage display
- PMID: 15963231
- PMCID: PMC1183247
- DOI: 10.1186/1477-5956-3-7
Cognate peptide-receptor ligand mapping by directed phage display
Abstract
Background: A rapid phage display method for the elucidation of cognate peptide specific ligand for receptors is described. The approach may be readily integrated into the interface of genomic and proteomic studies to identify biologically relevant ligands.
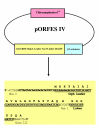
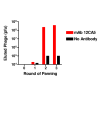
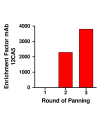
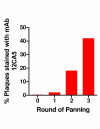
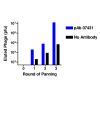
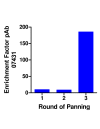
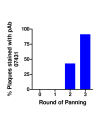
Methods: A gene fragment library from influenza coat protein haemagglutinin (HA) gene was constructed by treating HA cDNA with DNAse I to create 50-100 bp fragments. These fragments were cloned into plasmid pORFES IV and in-frame inserts were selected. These in-frame fragment inserts were subsequently cloned into a filamentous phage display vector JC-M13-88 for surface display as fusions to a synthetic copy of gene VIII. Two well characterized antibodies, mAb 12CA5 and pAb 07431, directed against distinct known regions of HA were used to pan the library.
Results: Two linear epitopes, HA peptide 112-126 and 162-173, recognized by mAb 12CA5 and pAb 07431, respectively, were identified as the cognate epitopes.
Conclusion: This approach is a useful alternative to conventional methods such as screening of overlapping synthetic peptide libraries or gene fragment expression libraries when searching for precise peptide protein interactions, and may be applied to functional proteomics.
Figures
Similar articles
-
Mapping of linear epitopes recognized by monoclonal antibodies with gene-fragment phage display libraries.Mol Gen Genet. 1995 Dec 10;249(4):425-31. doi: 10.1007/BF00287104. Mol Gen Genet. 1995. PMID: 8552047
-
Epitope mapping by phage display: random versus gene-fragment libraries.J Immunol Methods. 1997 Aug 7;206(1-2):43-52. doi: 10.1016/s0022-1759(97)00083-5. J Immunol Methods. 1997. PMID: 9328567
-
Mapping of hiv-1 Gag epitopes recognized by polyclonal antibodies using gene-fragment phage display system.Prep Biochem Biotechnol. 2001 May;31(2):185-200. doi: 10.1081/PB-100103383. Prep Biochem Biotechnol. 2001. PMID: 11426705
-
Phage display: concept, innovations, applications and future.Biotechnol Adv. 2010 Nov-Dec;28(6):849-58. doi: 10.1016/j.biotechadv.2010.07.004. Epub 2010 Jul 23. Biotechnol Adv. 2010. PMID: 20659548 Review.
-
Protein-protein interactions in hematology and phage display.Exp Hematol. 2001 Oct;29(10):1136-46. doi: 10.1016/s0301-472x(01)00693-2. Exp Hematol. 2001. PMID: 11602315 Review.
Cited by
-
Biomarker Discovery by ORFeome Phage Display.Methods Mol Biol. 2023;2702:543-561. doi: 10.1007/978-1-0716-3381-6_27. Methods Mol Biol. 2023. PMID: 37679638
-
Parameter estimate of signal transduction pathways.BMC Neurosci. 2006 Oct 30;7 Suppl 1(Suppl 1):S6. doi: 10.1186/1471-2202-7-S1-S6. BMC Neurosci. 2006. PMID: 17118160 Free PMC article. Review.
-
Proteomics technologies and challenges.Genomics Proteomics Bioinformatics. 2007 May;5(2):77-85. doi: 10.1016/S1672-0229(07)60018-7. Genomics Proteomics Bioinformatics. 2007. PMID: 17893073 Free PMC article. Review.
References
-
- Patel SD, Cope AP, Congia M, Chen TT, Kim E, Fugger L, Wherrett D, Sonderstrup-McDevitt G. Identification of immunodominant T cell epitopes of human glutamic acid decarboxylase 65 by using HLA-DR(alpha1*0101,beta1*0401) transgenic mice. Proc Natl Acad Sci U S A. 1997;94:8082–7. doi: 10.1073/pnas.94.15.8082. - DOI - PMC - PubMed
-
- Aoubala M, Douchet I, Bezzine S, Hirn M, Verger R, De Caro A. Immunological techniques for the characterization of digestive lipases. Methods Enzymol. 1997;286:126–49. - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

