The cotranscriptional assembly of snoRNPs controls the biosynthesis of H/ACA snoRNAs in Saccharomyces cerevisiae
- PMID: 15964797
- PMCID: PMC1156983
- DOI: 10.1128/MCB.25.13.5396-5403.2005
The cotranscriptional assembly of snoRNPs controls the biosynthesis of H/ACA snoRNAs in Saccharomyces cerevisiae
Abstract
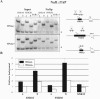
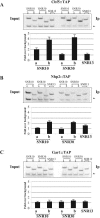
The carboxy-terminal domain (CTD) of RNA polymerase II large subunit acts as a platform to assemble the RNA processing machinery in a controlled way throughout the transcription cycle. In yeast, recent findings revealed a physical connection between phospho-CTD, generated by the Ctk1p kinase, and protein factors having a function in small nucleolar RNA (snoRNA) biogenesis. The snoRNAs represent a large family of polymerase II noncoding transcripts that are associated with highly conserved polypeptides to form stable ribonucleoprotein particles (snoRNPs). In this work, we have studied the biogenesis of the snoRNPs belonging to the box H/ACA class. We report that the assembly factor Naf1p and the core components Cbf5p and Nhp2p are recruited on H/ACA snoRNA genes very early during transcription. We also show that the cotranscriptional recruitment of Naf1p and Cbf5p is Ctk1p dependent and that Ctk1p and Cbf5p are required for preventing the readthrough into the snoRNA downstream genes. All these data suggest that proper cotranscriptional snoRNP assembly controls 3'-end formation of snoRNAs and, consequently, the release of a functional particle.
Figures



References
-
- Ahn, S. H., M. Kim, and S. Buratowski. 2004. Phosphorylation of serine 2 within the RNA polymerase II C-terminal domain couples transcription and 3′ end processing. Mol. Cell 13:67-76. - PubMed
-
- Corden, J. L. 1990. Tails of RNA polymerase II. Trends Biochem. Sci. 15:383-387. - PubMed
-
- Dez, C., A. Henras, B. Faucon, D. Lafontaine, M. Caizergues-Ferrer, and Y. Henry. 2001. Stable expression in yeast of the mature form of human telomerase RNA depends on its association with the box H/ACA small nucleolar RNP proteins Cbf5p, Nhp2p and Nop10p. Nucleic Acids Res. 29:598-603. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
