Generation and characterization of Rac3 knockout mice
- PMID: 15964829
- PMCID: PMC1156997
- DOI: 10.1128/MCB.25.13.5763-5776.2005
Generation and characterization of Rac3 knockout mice
Abstract
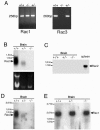
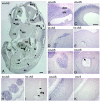
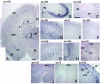
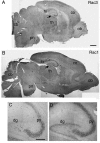
Rac proteins are members of the Rho family of GTPases involved in the regulation of actin dynamics. The three highly homologous Rac proteins in mammals are the ubiquitous Rac1, the hematopoiesis-specific Rac2, and the least-characterized Rac3. We show here that Rac3 mRNA is widely and specifically expressed in the developing nervous system, with highest concentration at embryonic day 13 in the dorsal root ganglia and ventral spinal cord. At postnatal day 7 Rac3 appears particularly abundant in populations of projection neurons in several regions of the brain, including the fifth layer of the cortex and the CA1-CA3 region of the hippocampus. We generated mice deleted for the Rac3 gene with the aim of analyzing the function of this GTPase in vivo. Rac3 knockout animals survive embryogenesis and show no obvious developmental defects. Interestingly, specific behavioral differences were detected in the Rac3-deficient animals, since motor coordination and motor learning on the rotarod was superior to that of their wild-type littermates. No obvious histological or immunohistological differences were observed at major sites of Rac3 expression. Our results indicate that, in vivo, Rac3 activity is not strictly required for normal development in utero but may be relevant to later events in the development of a functional nervous system.
Figures






References
-
- Abdel-Latif, D., M. Steward, D. L. Macdonald, G. A. Francis, M. C. Dinauer, and P. Lacy. 2004. Rac2 is critical for neutrophil primary granule exocytosis. Blood 104:832-839. - PubMed
-
- Abo, A., E. Pick, A. Hall, N. Totty, C. G. Teahan, and A. W. Segal. 1991. Activation of the NADPH oxidase involves the small GTP-binding protein p21rac1. Nature 353:668-670. - PubMed
-
- Ando, S., K. Kaibuchi, T. Sasaki, K. Hiraoka, T. Nishiyama, T. Mizuno, M. Asada, H. Nunoi, I. Matsuda, Y. Matsuura, P. Polakis, F. McCormick, and Y. Takai. 1992. Post-translational processing of Rac p21s is important both for their interaction with the GDP/GTP exchange proteins and for their activation of NADPH oxidase. J. Biol. Chem. 267:25709-25713. - PubMed
-
- Benvenuti, F., S. Hugues, M. Walmsley, S. Ruf, L. Fetler, M. Popoff, V. L. Tybulewicz, and S. Amigorena. 2004. Requirement of Rac1 and Rac2 expression by mature dendritic cells for T cell priming. Science 305:1150-1153. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
