Variable lymphocyte receptors in hagfish
- PMID: 15964979
- PMCID: PMC1166628
- DOI: 10.1073/pnas.0503792102
Variable lymphocyte receptors in hagfish
Abstract
A previously uncharacterized type of variable lymphocyte receptors (VLR) was identified recently in the Sea lamprey. This jawless vertebrate generates an extensive VLR repertoire through differential insertion of neighboring diverse leucine-rich repeat (LRR) cassettes into an incomplete germ-line VLR gene. We report here VLR homologs from two additional lamprey species and the presence of two types of VLR genes in hagfish, the only other order of contemporary jawless vertebrates. As in the Sea lamprey, the incomplete hagfish germ-line VLR-A and -B genes are modified in lymphocyte-like cells to generate highly diverse repertoires of VLR-A and -B proteins via a presently undetermined mechanism. This jawless-fish mode of VLR diversification starkly contrasts with the rearrangement of Ig V(D)J gene segments used by all jawed vertebrates to produce diverse repertoires of T and B lymphocyte antigen receptors. The development of two very different strategies for receptor diversification at the dawn of vertebrate evolution approximately 500 million years ago attests to the fitness value of a lymphocyte-based system of anticipatory immunity.
Figures

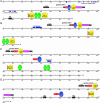

References
-
- Delarbre, C., Gallut, C, Barriel, V., Janvier, P. & Gachelin, G. (2002) Mol. Phylogenet. Evol. 22, 184-192. - PubMed
-
- Furlong, R. F. & Holland, P. W. (2002) Zool. Sci. 19, 593-599. - PubMed
-
- Takezaki, N., Figueroa, F., Zaleska-Rutczynska. Z. & Klein, J. (2003) Mol. Biol. Evol. 20, 287-292. - PubMed
-
- Zapata, A., Fange, R., Mattisson, A. & Villena, A. (1984) Cell Tissue Res. 235, 691-693. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

