The Plasmodium falciparum cysteine protease falcipain-2 captures its substrate, hemoglobin, via a unique motif
- PMID: 15964982
- PMCID: PMC1166610
- DOI: 10.1073/pnas.0502368102
The Plasmodium falciparum cysteine protease falcipain-2 captures its substrate, hemoglobin, via a unique motif
Abstract
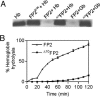
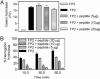
Falcipain-2 (FP2) is a papain family cysteine protease and important hemoglobinase of erythrocytic Plasmodium falciparum parasites. Inhibitors of FP2 block hemoglobin hydrolysis and parasite development, suggesting that this enzyme is a promising target for antimalarial chemotherapy. FP2 and related plasmodial cysteine proteases have an unusual 14-aa motif near the C terminus of the catalytic domain. Recent solution of the structure of FP2 showed this motif to form a beta-hairpin that is distant from the enzyme active site and protrudes out from the protein. To evaluate the function of this motif, we compared the activity of the wild-type enzyme with that of a mutant lacking 10 aa of the motif (Delta10FP2). Delta10FP2 had nearly identical activity to that of the wild-type enzyme against peptide substrates and the protein substrates casein and gelatin. However, Delta10FP2 demonstrated negligible activity against hemoglobin or globin. FP2 that was inhibited with trans-epoxysuccinyl-L-leucylamido-(4-guanidino)butane (FP2E-64) formed a complex with hemoglobin, but Delta10FP2E-64 did not, indicating that the motif mediates binding to hemoglobin independent of the active site. A peptide encoding the motif blocked hemoglobin hydrolysis, but not the hydrolysis of casein. Kinetics for the inhibition of Delta10FP2 were very similar to those for FP2 with peptidyl and protein inhibitors, but Delta10FP2 was poorly inhibited by the inhibitory prodomain of FP2. Our results indicate that FP2 utilizes an unusual motif for two specific functions, interaction with hemoglobin, its natural substrate, and interaction with the prodomain, its natural inhibitor.
Figures

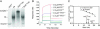
Similar articles
-
Structural basis for unique mechanisms of folding and hemoglobin binding by a malarial protease.Proc Natl Acad Sci U S A. 2006 Aug 1;103(31):11503-8. doi: 10.1073/pnas.0600489103. Epub 2006 Jul 24. Proc Natl Acad Sci U S A. 2006. PMID: 16864794 Free PMC article.
-
Independent intramolecular mediators of folding, activity, and inhibition for the Plasmodium falciparum cysteine protease falcipain-2.J Biol Chem. 2004 Jan 30;279(5):3484-91. doi: 10.1074/jbc.M310536200. Epub 2003 Nov 18. J Biol Chem. 2004. PMID: 14625277
-
Hemoglobin cleavage site-specificity of the Plasmodium falciparum cysteine proteases falcipain-2 and falcipain-3.PLoS One. 2009;4(4):e5156. doi: 10.1371/journal.pone.0005156. Epub 2009 Apr 9. PLoS One. 2009. PMID: 19357776 Free PMC article.
-
Cysteine proteases of malaria parasites.Int J Parasitol. 2004 Dec;34(13-14):1489-99. doi: 10.1016/j.ijpara.2004.10.003. Int J Parasitol. 2004. PMID: 15582526 Review.
-
Falcipains and other cysteine proteases of malaria parasites.Adv Exp Med Biol. 2011;712:30-48. doi: 10.1007/978-1-4419-8414-2_3. Adv Exp Med Biol. 2011. PMID: 21660657 Review.
Cited by
-
Enantioselective Synthesis of γ-Phenyl-γ-amino Vinyl Phosphonates and Sulfones and Their Application to the Synthesis of Novel Highly Potent Antimalarials.ACS Omega. 2020 Nov 2;5(45):29025-29037. doi: 10.1021/acsomega.0c03470. eCollection 2020 Nov 17. ACS Omega. 2020. PMID: 33225134 Free PMC article.
-
Falstatin, a cysteine protease inhibitor of Plasmodium falciparum, facilitates erythrocyte invasion.PLoS Pathog. 2006 Nov;2(11):e117. doi: 10.1371/journal.ppat.0020117. PLoS Pathog. 2006. PMID: 17083274 Free PMC article.
-
Methyl-methoxylpyrrolinone and flavinium nucleus binding signatures on falcipain-2 active site.J Mol Model. 2014 Aug;20(8):2386. doi: 10.1007/s00894-014-2386-2. Epub 2014 Aug 6. J Mol Model. 2014. PMID: 25096811
-
The Ionic and hydrophobic interactions are required for the auto activation of cysteine proteases of Plasmodium falciparum.PLoS One. 2012;7(10):e47227. doi: 10.1371/journal.pone.0047227. Epub 2012 Oct 16. PLoS One. 2012. PMID: 23077573 Free PMC article.
-
Engineering Nucleotide Specificity of Succinyl-CoA Synthetase in Blastocystis: The Emerging Role of Gatekeeper Residues.Biochemistry. 2017 Jan 24;56(3):534-542. doi: 10.1021/acs.biochem.6b00098. Epub 2017 Jan 11. Biochemistry. 2017. PMID: 27478903 Free PMC article.
References
-
- Breman, J. G. (2001) Am. J. Trop. Med. Hyg. 64, 1-11. - PubMed
-
- Rosenthal, P. J. (2003) J. Exp. Biol. 206, 3735-3744. - PubMed
-
- Francis, S. E., Sullivan, D. J. & Goldberg, D. E. (1997) Annu. Rev. Microbiol. 51, 97-123. - PubMed
-
- Lew, V. L., Tiffert, T. & Ginsburg, H. (2003) Blood 101, 4189-4194. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

