Tsix transcription across the Xist gene alters chromatin conformation without affecting Xist transcription: implications for X-chromosome inactivation
- PMID: 15964997
- PMCID: PMC1151664
- DOI: 10.1101/gad.341105
Tsix transcription across the Xist gene alters chromatin conformation without affecting Xist transcription: implications for X-chromosome inactivation
Abstract
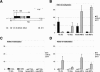
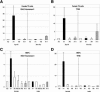
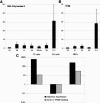
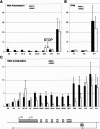
X-chromosome inactivation (XCI) is highly dynamic during early mouse embryogenesis and strictly depends on the Xist noncoding RNA. The regulation of Xist and its antisense partner Tsix remains however poorly understood. We provide here the first evidence of transcriptional control of Xist expression. We show that RNA polymerase II (RNAPolII) preinitiation complex recruitment and H3 Lys 4 (H3-K4) methylation at the Xist promoter form the basis of the Xist expression profiles that drives both imprinted and random XCI. In embryonic stem (ES) cells, which are derived from the inner cell mass where imprinted XCI is reversed and both Xs are active, we show that Xist is repressed at the level of preinitiation complex (PIC) recruitment. We further demonstrate that Tsix, although highly transcribed in ES cells, is not itself responsible for the transcriptional down-regulation of Xist. Rather, Tsix induces efficient H3-K4 methylation over the entire Xist/Tsix unit. We suggest that chromatin remodeling of the Xist locus induced by biallelic Tsix transcription renders both Xist loci epigenetically equivalent and equally competent for transcription. In this model, Tsix, by resetting the epigenetic state of the Xist/Tsix locus, mediates the transition from imprinted to random XCI.
Figures
Similar articles
-
Tsix-mediated epigenetic switch of a CTCF-flanked region of the Xist promoter determines the Xist transcription program.Genes Dev. 2006 Oct 15;20(20):2787-92. doi: 10.1101/gad.389006. Genes Dev. 2006. PMID: 17043308 Free PMC article.
-
Tsix silences Xist through modification of chromatin structure.Dev Cell. 2005 Jul;9(1):159-65. doi: 10.1016/j.devcel.2005.05.015. Dev Cell. 2005. PMID: 15992549
-
Crucial role of antisense transcription across the Xist promoter in Tsix-mediated Xist chromatin modification.Development. 2008 Jan;135(2):227-35. doi: 10.1242/dev.008490. Epub 2007 Dec 5. Development. 2008. PMID: 18057104
-
X-tra! X-tra! News from the mouse X chromosome.Dev Biol. 2006 Oct 15;298(2):344-53. doi: 10.1016/j.ydbio.2006.07.011. Epub 2006 Jul 15. Dev Biol. 2006. PMID: 16916508 Review.
-
An embryonic story: analysis of the gene regulative network controlling Xist expression in mouse embryonic stem cells.Bioessays. 2010 Jul;32(7):581-8. doi: 10.1002/bies.201000019. Bioessays. 2010. PMID: 20544740 Review.
Cited by
-
The X as model for RNA's niche in epigenomic regulation.Cold Spring Harb Perspect Biol. 2010 Sep;2(9):a003749. doi: 10.1101/cshperspect.a003749. Epub 2010 Mar 31. Cold Spring Harb Perspect Biol. 2010. PMID: 20739414 Free PMC article. Review.
-
Telomeric RNAs mark sex chromosomes in stem cells.Genetics. 2009 Jul;182(3):685-98. doi: 10.1534/genetics.109.103093. Epub 2009 Apr 20. Genetics. 2009. PMID: 19380904 Free PMC article.
-
Higher order chromatin structure at the X-inactivation center via looping DNA.Dev Biol. 2008 Jul 15;319(2):416-25. doi: 10.1016/j.ydbio.2008.04.010. Epub 2008 Apr 18. Dev Biol. 2008. PMID: 18501343 Free PMC article.
-
A regulatory potential of the Xist gene promoter in vole M. rossiaemeridionalis.PLoS One. 2012;7(5):e33994. doi: 10.1371/journal.pone.0033994. Epub 2012 May 11. PLoS One. 2012. PMID: 22606223 Free PMC article.
-
Clusters of internally primed transcripts reveal novel long noncoding RNAs.PLoS Genet. 2006 Apr;2(4):e37. doi: 10.1371/journal.pgen.0020037. Epub 2006 Apr 28. PLoS Genet. 2006. PMID: 16683026 Free PMC article.
References
-
- Borsani B., Tonlorenzi, R., Simmler, M.-C., Dandolo, L., Arnaud, D., Capra, V., Grompe, M., Pizzuti, A., Muzni, D., Lawrence, C., et al. 1991. Characterization of a murine gene expressed from the inactive X chromosome. Nature 351: 325-329. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
