X-linked genes evolve higher codon bias in Drosophila and Caenorhabditis
- PMID: 15965246
- PMCID: PMC1456507
- DOI: 10.1534/genetics.105.043497
X-linked genes evolve higher codon bias in Drosophila and Caenorhabditis
Abstract
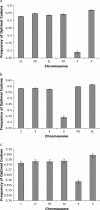
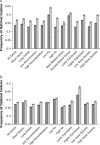
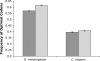
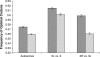
Comparing patterns of molecular evolution between autosomes and sex chromosomes (such as X and W chromosomes) can provide insight into the forces underlying genome evolution. Here we investigate patterns of codon bias evolution on the X chromosome and autosomes in Drosophila and Caenorhabditis. We demonstrate that X-linked genes have significantly higher codon bias compared to autosomal genes in both Drosophila and Caenorhabditis. Furthermore, genes that become X-linked evolve higher codon bias gradually, over tens of millions of years. We provide several lines of evidence that this elevation in codon bias is due exclusively to their chromosomal location and not to any other property of X-linked genes. We present two possible explanations for these observations. One possibility is that natural selection is more efficient on the X chromosome due to effective haploidy of the X chromosomes in males and persistently low effective numbers of reproducing males compared to that of females. Alternatively, X-linked genes might experience stronger natural selection for higher codon bias as a result of maladaptive reduction of their dosage engendered by the loss of the Y-linked homologs.
Figures
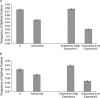
Similar articles
-
Codon bias and noncoding GC content correlate negatively with recombination rate on the Drosophila X chromosome.J Mol Evol. 2005 Sep;61(3):315-24. doi: 10.1007/s00239-004-0287-1. Epub 2005 Jul 21. J Mol Evol. 2005. PMID: 16044248
-
Hill-Robertson interference is a minor determinant of variations in codon bias across Drosophila melanogaster and Caenorhabditis elegans genomes.Mol Biol Evol. 2002 Sep;19(9):1399-406. doi: 10.1093/oxfordjournals.molbev.a004203. Mol Biol Evol. 2002. PMID: 12200468
-
Codon usage bias and effective population sizes on the X chromosome versus the autosomes in Drosophila melanogaster.Mol Biol Evol. 2013 Apr;30(4):811-23. doi: 10.1093/molbev/mss222. Epub 2012 Nov 29. Mol Biol Evol. 2013. PMID: 23204387 Free PMC article.
-
Selection on codon bias.Annu Rev Genet. 2008;42:287-99. doi: 10.1146/annurev.genet.42.110807.091442. Annu Rev Genet. 2008. PMID: 18983258 Review.
-
Non-random autosome segregation: a stepping stone for the evolution of sex chromosome complexes? Sex-biased transmission of autosomes could facilitate the spread of antagonistic alleles, and generate sex-chromosome systems with multiple X or Y chromosomes.Bioessays. 2011 Feb;33(2):111-4. doi: 10.1002/bies.201000106. Bioessays. 2011. PMID: 21154781 Review.
Cited by
-
Contrasting Levels of Molecular Evolution on the Mouse X Chromosome.Genetics. 2016 Aug;203(4):1841-57. doi: 10.1534/genetics.116.186825. Epub 2016 Jun 17. Genetics. 2016. PMID: 27317678 Free PMC article.
-
Recombination rates may affect the ratio of X to autosomal noncoding polymorphism in African populations of Drosophila melanogaster.Genetics. 2009 Apr;181(4):1699-701; author reply 1703. doi: 10.1534/genetics.108.098004. Epub 2009 Feb 2. Genetics. 2009. PMID: 19189953 Free PMC article. No abstract available.
-
Patterns of DNA-sequence divergence between Drosophila miranda and D. pseudoobscura.J Mol Evol. 2009 Dec;69(6):601-11. doi: 10.1007/s00239-009-9298-2. Epub 2009 Oct 27. J Mol Evol. 2009. PMID: 19859648
-
An excess of gene expression divergence on the X chromosome in Drosophila embryos: implications for the faster-X hypothesis.PLoS Genet. 2012;8(12):e1003200. doi: 10.1371/journal.pgen.1003200. Epub 2012 Dec 27. PLoS Genet. 2012. PMID: 23300473 Free PMC article.
-
Recent and Long-Term Selection Across Synonymous Sites in Drosophila ananassae.J Mol Evol. 2016 Aug;83(1-2):50-60. doi: 10.1007/s00239-016-9753-9. Epub 2016 Aug 1. J Mol Evol. 2016. PMID: 27481397 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

