Genetic instability induced by overexpression of DNA ligase I in budding yeast
- PMID: 15965249
- PMCID: PMC1456761
- DOI: 10.1534/genetics.105.042861
Genetic instability induced by overexpression of DNA ligase I in budding yeast
Abstract
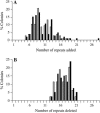
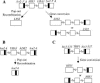
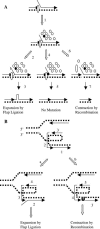
Recombination and microsatellite mutation in humans contribute to disorders including cancer and trinucleotide repeat (TNR) disease. TNR expansions in wild-type yeast may arise by flap ligation during lagging-strand replication. Here we show that overexpression of DNA ligase I (CDC9) increases the rates of TNR expansion, of TNR contraction, and of mitotic recombination. Surprisingly, this effect is observed with catalytically inactive forms of Cdc9p protein, but only if they possess a functional PCNA-binding site. Furthermore, in vitro analysis indicates that the interaction of PCNA with Cdc9p and Rad27p (Fen1) is mutually exclusive. Together our genetic and biochemical analysis suggests that, although DNA ligase I seals DNA nicks during replication, repair, and recombination, higher than normal levels can yield genetic instability by disrupting the normal interplay of PCNA with other proteins such as Fen1.
Figures


References
-
- Ayyagari, R., X. V. Gomes, D. A. Gordenin and P. M. Burgers, 2003. Okazaki fragment maturation in yeast. I. Distribution of functions between FEN1 and DNA2. J. Biol. Chem. 278: 1618–1625. - PubMed
-
- Bae, S. H., and Y. S. Seo, 2000. Characterization of the enzymatic properties of the yeast dna2 helicase/endonuclease suggests a new model for Okazaki fragment processing. J. Biol. Chem. 275: 38022–38031. - PubMed
-
- Bae, S. H., K. H. Bae, J. A. Kim and Y. S. Seo, 2001. RPA governs endonuclease switching during processing of Okazaki fragments in eukaryotes. Nature 412: 456–461. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

