A segmental deletion series generated by sister-chromatid transposition of Ac transposable elements in maize
- PMID: 15965263
- PMCID: PMC1456524
- DOI: 10.1534/genetics.104.035576
A segmental deletion series generated by sister-chromatid transposition of Ac transposable elements in maize
Abstract
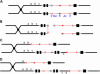
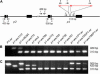
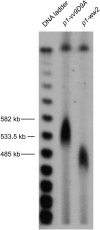
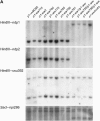
Certain configurations of maize Ac/Ds transposon termini can undergo alternative transposition reactions leading to chromosome breakage and various types of stable chromosome rearrangements. Here, we show that a particular allele of the maize p1 gene containing an intact Ac element and a nearby terminally deleted Ac element (fAc) can undergo sister-chromatid transposition (SCT) reactions that generate large flanking deletions. Among 35 deletions characterized, all begin at the Ac termini in the p1 gene and extend to various flanking sites proximal to p1. The deletions range in size from the smallest of 12,567 bp to the largest of >4.6 cM; >80% of the deletions removed the p2 gene, a paralog of p1 located approximately 60 kb from p1 in the p1-vv allele and its derivatives. Sequencing of representative cases shows that the deletions have precise junctions between the transposon termini and the flanking genomic sequences. These results show that SCT events can efficiently generate interstitial deletions that are useful for in vivo dissection of local genome regions and for the rapid correlation of genetic and physical maps. Finally, we discuss evidence suggesting that deletions induced by alternative transposition reactions can occur at other genomic loci, indicating that this mechanism may have had a significant impact on genome evolution.
Figures


References
-
- Anderson, P. A., P. A. Okubara, R. Arroyo-Garcia, B. C. Meyers and R. W. Michelmore, 1996. Molecular analysis of irradiation-induced and spontaneous deletion mutants at a disease resistance locus in Lactuca sativa. Mol. Gen. Genet. 251: 316–325. - PubMed
-
- Bayley, C. C., M. Morgan, E. C. Dale and D. W. Ow, 1992. Exchange of gene activity in transgenic plants catalyzed by the Cre-lox site-specific recombination system. Plant Mol. Biol. 18: 353–361. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

