An integrative genomics approach to infer causal associations between gene expression and disease
- PMID: 15965475
- PMCID: PMC2841396
- DOI: 10.1038/ng1589
An integrative genomics approach to infer causal associations between gene expression and disease
Abstract
A key goal of biomedical research is to elucidate the complex network of gene interactions underlying complex traits such as common human diseases. Here we detail a multistep procedure for identifying potential key drivers of complex traits that integrates DNA-variation and gene-expression data with other complex trait data in segregating mouse populations. Ordering gene expression traits relative to one another and relative to other complex traits is achieved by systematically testing whether variations in DNA that lead to variations in relative transcript abundances statistically support an independent, causative or reactive function relative to the complex traits under consideration. We show that this approach can predict transcriptional responses to single gene-perturbation experiments using gene-expression data in the context of a segregating mouse population. We also demonstrate the utility of this approach by identifying and experimentally validating the involvement of three new genes in susceptibility to obesity.
Conflict of interest statement
The authors declare that they have no competing financial interests.
Figures
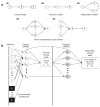


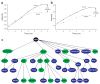
References
-
- Hughes TR, et al. Functional discovery via a compendium of expression profiles. Cell. 2000;102:109–126. - PubMed
-
- Karp CL, et al. Identification of complement factor 5 as a susceptibility locus for experimental allergic asthma. Nat Immunol. 2000;1:221–226. - PubMed
-
- Schadt EE, et al. Genetics of gene expression surveyed in maize, mouse and man. Nature. 2003;422:297–302. - PubMed
-
- Johnson JM, et al. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science. 2003;302:2141–2144. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

