H-NS is a part of a thermally controlled mechanism for bacterial gene regulation
- PMID: 15966862
- PMCID: PMC1276917
- DOI: 10.1042/BJ20050453
H-NS is a part of a thermally controlled mechanism for bacterial gene regulation
Abstract
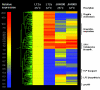
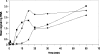
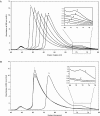
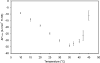
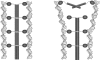
Temperature is a primary environmental stress to which micro-organisms must be able to adapt and respond rapidly. Whereas some bacteria are restricted to specific niches and have limited abilities to survive changes in their environment, others, such as members of the Enterobacteriaceae, can withstand wide fluctuations in temperature. In addition to regulating cellular physiology, pathogenic bacteria use temperature as a cue for activating virulence gene expression. This work confirms that the nucleoid-associated protein H-NS (histone-like nucleoid structuring protein) is an essential component in thermoregulation of Salmonella. On increasing the temperature from 25 to 37 degrees C, more than 200 genes from Salmonella enterica serovar Typhimurium showed H-NS-dependent up-regulation. The thermal activation of gene expression is extremely rapid and change in temperature affects the DNA-binding properties of H-NS. The reduction in gene repression brought about by the increase in temperature is concomitant with a conformational change in the protein, resulting in the decrease in size of high-order oligomers and the appearance of increasing concentrations of discrete dimers of H-NS. The present study addresses one of the key complex mechanisms by which H-NS regulates gene expression.
Figures

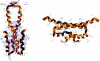
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

