Functional analysis of the Burkholderia cenocepacia ZmpA metalloprotease
- PMID: 15968051
- PMCID: PMC1151788
- DOI: 10.1128/JB.187.13.4421-4429.2005
Functional analysis of the Burkholderia cenocepacia ZmpA metalloprotease
Abstract
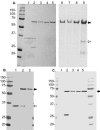
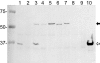
Burkholderia cenocepacia ZmpA is expressed as a preproenzyme typical of thermolysin-like proteases such as Pseudomonas aeruginosa LasB and Bacillus thermoproteolyticus thermolysin. The zmpA gene was expressed using the pPRO-EXHTa His(6) tag expression system, which incorporates a six-His tag at the N-terminal end of the protein, and recombinant ZmpA was purified using Ni-nitrilotriacetic acid affinity chromatography. Upon refolding of the recombinant His(6)-pre-pro-ZmpA (62 kDa), the fusion protein was autoproteolytically cleaved into 36-kDa (mature ZmpA) and 27-kDa peptides. Site-directed mutagenesis was employed to infer the identity of the active site residues of ZmpA and to confirm that the enzyme undergoes autoproteolytic cleavage. Oligonucleotide mutagenesis was used to replace H(465) with G(465) or A(465), E(377) with A(377) or D(377), or H(380) with P(380) or A(380). Mutagenesis of H(465), E(377), or H(380) resulted in the loss of both autocatalytic activity and proteolytic activity. ZmpA with either substitution in H(380) was not detectable in B. cenocepacia cell extracts. The activity of the recombinant ZmpA was inhibited by EDTA and 1,10 phenanthroline, indicating that it is a zinc metalloprotease. ZmpA, however, was not inhibited by phosphoramidon, a classical inhibitor of the thermolysin-like proteases. The refolded mature ZmpA enzyme was proteolytically active against various substrates including hide powder azure, type IV collagen, fibronectin, neutrophil alpha-1 proteinase inhibitor, alpha(2)-macroglobulin, and gamma interferon, suggesting that B. cenocepacia ZmpA may cause direct tissue damage to the host or damage to host tissues through a modulation of the host's immune system.
Figures
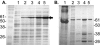

Similar articles
-
Distribution and expression of the ZmpA metalloprotease in the Burkholderia cepacia complex.J Bacteriol. 2005 Dec;187(24):8247-55. doi: 10.1128/JB.187.24.8247-8255.2005. J Bacteriol. 2005. PMID: 16321929 Free PMC article.
-
Burkholderia cenocepacia ZmpB is a broad-specificity zinc metalloprotease involved in virulence.Infect Immun. 2006 Jul;74(7):4083-93. doi: 10.1128/IAI.00297-06. Infect Immun. 2006. PMID: 16790782 Free PMC article.
-
Burkholderia cenocepacia zinc metalloproteases influence resistance to antimicrobial peptides.Microbiology (Reading). 2009 Sep;155(Pt 9):2818-2825. doi: 10.1099/mic.0.028969-0. Epub 2009 Jun 18. Microbiology (Reading). 2009. PMID: 19542010
-
An extracellular zinc metalloprotease gene of Burkholderia cepacia.Microbiology (Reading). 2003 Aug;149(Pt 8):2263-2271. doi: 10.1099/mic.0.26243-0. Microbiology (Reading). 2003. PMID: 12904566
-
A substitution at His-120 in the LasA protease of Pseudomonas aeruginosa blocks enzymatic activity without affecting propeptide processing or extracellular secretion.J Bacteriol. 1996 Nov;178(22):6608-17. doi: 10.1128/jb.178.22.6608-6617.1996. J Bacteriol. 1996. PMID: 8932318 Free PMC article.
Cited by
-
Identification of Burkholderia cenocepacia strain H111 virulence factors using nonmammalian infection hosts.Infect Immun. 2013 Jan;81(1):143-53. doi: 10.1128/IAI.00768-12. Epub 2012 Oct 22. Infect Immun. 2013. PMID: 23090963 Free PMC article.
-
The metalloprotease of Listeria monocytogenes is activated by intramolecular autocatalysis.J Bacteriol. 2008 Jan;190(1):107-11. doi: 10.1128/JB.00852-07. Epub 2007 Oct 26. J Bacteriol. 2008. PMID: 17965168 Free PMC article.
-
Distribution and expression of the ZmpA metalloprotease in the Burkholderia cepacia complex.J Bacteriol. 2005 Dec;187(24):8247-55. doi: 10.1128/JB.187.24.8247-8255.2005. J Bacteriol. 2005. PMID: 16321929 Free PMC article.
-
Interferon-gamma-activated macrophages infected with Burkholderia cenocepacia process and present bacterial antigens to T-cells by class I and II major histocompatibility complex molecules.Emerg Microbes Infect. 2020 Dec;9(1):2000-2012. doi: 10.1080/22221751.2020.1818632. Emerg Microbes Infect. 2020. PMID: 32873215 Free PMC article.
-
Iron and zinc exploitation during bacterial pathogenesis.Metallomics. 2015 Dec;7(12):1541-54. doi: 10.1039/c5mt00170f. Epub 2015 Oct 26. Metallomics. 2015. PMID: 26497057 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

