Complete genome sequence and analysis of the multiresistant nosocomial pathogen Corynebacterium jeikeium K411, a lipid-requiring bacterium of the human skin flora
- PMID: 15968079
- PMCID: PMC1151758
- DOI: 10.1128/JB.187.13.4671-4682.2005
Complete genome sequence and analysis of the multiresistant nosocomial pathogen Corynebacterium jeikeium K411, a lipid-requiring bacterium of the human skin flora
Abstract
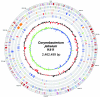
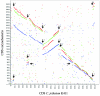
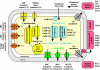
Corynebacterium jeikeium is a "lipophilic" and multidrug-resistant bacterial species of the human skin flora that has been recognized with increasing frequency as a serious nosocomial pathogen. Here we report the genome sequence of the clinical isolate C. jeikeium K411, which was initially recovered from the axilla of a bone marrow transplant patient. The genome of C. jeikeium K411 consists of a circular chromosome of 2,462,499 bp and the 14,323-bp bacteriocin-producing plasmid pKW4. The chromosome of C. jeikeium K411 contains 2,104 predicted coding sequences, 52% of which were considered to be orthologous with genes in the Corynebacterium glutamicum, Corynebacterium efficiens, and Corynebacterium diphtheriae genomes. These genes apparently represent the chromosomal backbone that is conserved between the four corynebacteria. Among the genes that lack an ortholog in the known corynebacterial genomes, many are located close to transposable elements or revealed an atypical G+C content, indicating that horizontal gene transfer played an important role in the acquisition of genes involved in iron and manganese homeostasis, in multidrug resistance, in bacterium-host interaction, and in virulence. Metabolic analyses of the genome sequence indicated that the "lipophilic" phenotype of C. jeikeium most likely originates from the absence of fatty acid synthase and thus represents a fatty acid auxotrophy. Accordingly, both the complete gene repertoire and the deduced lifestyle of C. jeikeium K411 largely reflect the strict dependence of growth on the presence of exogenous fatty acids. The predicted virulence factors of C. jeikeium K411 are apparently involved in ensuring the availability of exogenous fatty acids by damaging the host tissue.
Figures
References
-
- Agranoff, D. D., and S. Krishna. 1998. Metal ion homeostasis and intracellular parasitism. Mol. Microbiol. 28:403-412. - PubMed
-
- Andrews, S. C., A. K. Robinson, and F. Rodríguez-Quiñones. 2003. Bacterial iron homeostasis. FEMS Microbiol. Rev. 27:215-237. - PubMed
-
- Balci, I., F. Eks̨i, and A. Bayram. 2002. Coryneform bacteria isolated from blood cultures and their antibiotic susceptibilities. J. Int. Med. Res. 30:422-427. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

