UDP-glucose dehydrogenases of maize: a role in cell wall pentose biosynthesis
- PMID: 15969652
- PMCID: PMC1276940
- DOI: 10.1042/BJ20050800
UDP-glucose dehydrogenases of maize: a role in cell wall pentose biosynthesis
Abstract
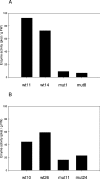
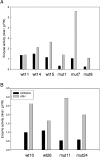
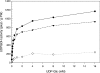
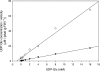
UDPGDH (UDP-D-glucose dehydrogenase) oxidizes UDP-Glc (UDP-D-glucose) to UDP-GlcA (UDP-D-glucuronate), the precursor of UDP-D-xylose and UDP-L-arabinose, major cell wall polysaccharide precursors. Maize (Zea mays L.) has at least two putative UDPGDH genes (A and B), according to sequence similarity to a soya bean UDPGDH gene. The predicted maize amino acid sequences have 95% similarity to that of soya bean. Maize mutants with a Mu-element insertion in UDPGDH-A or UDPGDH-B were isolated (udpgdh-A1 and udpgdh-B1 respectively) and studied for changes in wall polysaccharide biosynthesis. The udpgdh-A1 and udpgdh-B1 homozygotes showed no visible phenotype but exhibited 90 and 60-70% less UDPGDH activity respectively than wild-types in a radiochemical assay with 30 microM UDP-glucose. Ethanol dehydrogenase (ADH) activity varied independently of UDPGDH activity, supporting the hypothesis that ADH and UDPGDH activities are due to different enzymes in maize. When extracts from wild-types and udpgdh-A1 homozygotes were assayed with increasing concentrations of UDP-Glc, at least two isoforms of UDPGDH were detected, having K(m) values of approx. 380 and 950 microM for UDP-Glc. Leaf and stem non-cellulosic polysaccharides had lower Ara/Gal and Xyl/Gal ratios in udpgdh-A1 homozygotes than in wild-types, whereas udpgdh-B1 homozygotes exhibited more variability among individual plants, suggesting that UDPGDH-A activity has a more important role than UDPGDH-B in UDP-GlcA synthesis. The fact that mutation of a UDPGDH gene interferes with polysaccharide synthesis suggests a greater importance for the sugar nucleotide oxidation pathway than for the myo-inositol pathway in UDP-GlcA biosynthesis during post-germinative growth of maize.
Figures


References
-
- Feingold D. S., Avigad G. Sugar nucleotide transformations in plants. In: Preiss J., editor. The Biochemistry of Plants, A Comprehensive Treatise, vol. 3. New York: Academic Press; 1980. pp. 101–170.
-
- Mohnen D. Biosynthesis of pectins. In: Seymor G. B., Knox J. P., editors. Pectins and their Manipulation. Oxford, U.K.: Blackwell Publishing/CRC Press; 2002. pp. 52–98.
-
- Seifert G. J. Nucleotide sugar interconversions and cell wall biosynthesis: how to bring the inside to the outside. Curr. Opin. Plant Biol. 2004;7:277–284. - PubMed
-
- Carpita N. C. Structure and biogenesis of the cell walls of grasses. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1996;47:445–476. - PubMed
-
- Loewus F., Chen M. S., Loewus M. W. The myo-inositol oxidation pathway to cell wall polysaccharides. In: Loewus F., editor. Biogenesis of Plant Cell Wall Polysaccharides. New York: Academic Press; 1973. pp. 1–27.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

