Analysis of xylem sap proteins from Brassica napus
- PMID: 15969751
- PMCID: PMC1182380
- DOI: 10.1186/1471-2229-5-11
Analysis of xylem sap proteins from Brassica napus
Abstract
Background: Substance transport in higher land plants is mediated by vascular bundles, consisting of phloem and xylem strands that interconnect all plant organs. While the phloem mainly allocates photoassimilates, the role of the xylem is the transport of water and inorganic nutrients from roots to all aerial plant parts. Only recently it was noticed that in addition to mineral salts, xylem sap contains organic nutrients and even proteins. Although these proteins might have important impact on the performance of above-ground organs, only a few of them have been identified so far and their physiological functions are still unclear.
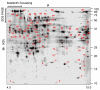
Results: We used root-pressure xylem exudate, collected from cut Brassica napus stems, to extract total proteins. These protein preparations were then separated by high-resolution two-dimensional gel electrophoresis (2-DE). After individual tryptic digests of the most abundant coomassie-stained protein spots, partial peptide sequence information was deduced from tandem mass spectrometric (MS/MS) fragmentation spectra and subsequently used for protein identifications by database searches. This approach resulted in the identification of 69 proteins. These identifications include different proteins potentially involved in defence-related reactions and cell wall metabolism.
Conclusion: This study provides a comprehensive overview of the most abundant proteins present in xylem sap of Brassica napus. A number of 69 proteins could be identified from which many previously were not known to be localized to this compartment in any other plant species. Since Brassica napus, a close relative of the fully sequenced model plant Arabidopsis thaliana, was used as the experimental system, our results provide a large number of candidate proteins for directed molecular and biochemical analyses of the physiological functions of the xylem under different environmental and developmental conditions. This approach will allow exploiting many of the already established functional genomic resources, like i.e. the large mutant collections, that are available for Arabidopsis.
Figures

References
-
- Schurr U, Schulze ED. The concentration of xylem sap constituents in root exudate, and in sap from intact, transpiring castor bean plants (Ricinus communis L.). Plant, Cell and Environment. 1995;18:409–420.
-
- Zornoza P, Gonzalez M, Serrano S, Carpena O. Intervarietal differences in xylem exudate composition and growth under contrasting forms of N supply in cucumber. Plant and Soil. 1996;178:311–317. doi: 10.1007/BF00011597. - DOI
-
- Satoh S, Kuroha T, Wakahoi T, Inouye Y. Inhibition of the formation of adventitious roots on cucumber hypocotyls by the fractions and methoxybenzylglutamine from xylem sap of squash root. Journal of Plant Research. 1998;111:541–546.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

