Quantitative trait loci affecting the difference in pigmentation between Drosophila yakuba and D. santomea
- PMID: 15972457
- PMCID: PMC1456512
- DOI: 10.1534/genetics.105.044412
Quantitative trait loci affecting the difference in pigmentation between Drosophila yakuba and D. santomea
Abstract
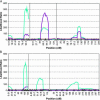
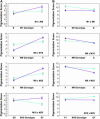
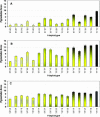
Using quantitative trait locus (QTL) mapping, we studied the genetic basis of the difference in pigmentation between two sister species of Drosophila: Drosophila yakuba, which, like other members of the D. melanogaster subgroup, shows heavy black pigmentation on the abdomen of males and females, and D. santomea, an endemic to the African island of São Tomé, which has virtually no pigmentation. Here we mapped four QTL with large effects on this interspecific difference in pigmentation: two on the X chromosome and one each on the second and third chromosomes. The same four QTL were detected in male hybrids in the backcrosses to both D. santomea and D. yakuba and in the female D. yakuba backcross hybrids. All four QTL exhibited strong epistatic interactions in male backcross hybrids, but only one pair of QTL interacted in females from the backcross to D. yabuka. All QTL from each species affected pigmentation in the same direction, consistent with adaptive evolution driven by directional natural selection. The regions delimited by the QTL included many positional candidate loci in the pigmentation pathway, including genes affecting catecholamine biosynthesis, melanization of the cuticle, and many additional pleiotropic effects.
Figures

References
-
- Ashburner, M., 1989. Drosophila: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
-
- Baier, A., B. Wittek and B. Brembs, 2002. Drosophila as a new model organism for the neurobiology of aggression? J. Exp. Biol. 205: 1233–1240. - PubMed
-
- Basten, C. J., B. S. Weir and Z-B. Zeng, 1999. QTL Cartographer, Version 1.13. Department of Statistics, North Carolina State University, Raleigh, NC.
-
- Blenau, W., and A. Baumann, 2001. Molecular and pharmacological properties of insect bioamine receptors: lessons from Drosophila melanogaster and Apis mellifera. Arch. Insect Biochem. Phys. 48: 13–38. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

