A genetic linkage map for the tiger pufferfish, Takifugu rubripes
- PMID: 15972462
- PMCID: PMC1456513
- DOI: 10.1534/genetics.105.042051
A genetic linkage map for the tiger pufferfish, Takifugu rubripes
Abstract
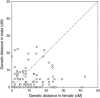
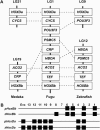
The compact genome of the tiger pufferfish, Takifugu rubripes (fugu), has been sequenced to the "draft" level and annotated to identify all the genes. However, the assembly of the draft genome sequence is highly fragmented due to the lack of a genetic or a physical map. To determine the long-range linkage relationship of the sequences, we have constructed the first genetic linkage map for fugu. The maps for the male and female spanning 697.1 and 1213.5 cM, respectively, were arranged into 22 linkage groups by markers heterozygous in both parents. The resulting map consists of 200 microsatellite loci physically linked to genome sequences spanning approximately 39 Mb in total. Comparisons of the genome maps of fugu, other teleosts, and mammals suggest that syntenic relationship is more conserved in the teleost lineage than in the mammalian lineage. Map comparisons also show a pufferfish lineage-specific rearrangement of the genome resulting in colocalization of two Hox gene clusters in one linkage group. This map provides a foundation for development of a complete physical map, a basis for comparison of long-range linkage of genes with other vertebrates, and a resource for mapping loci responsible for phenotypic differences among Takifugu species.
Figures




References
-
- Amores, A., A. Force, Y. L. Yan, L. Joly, C. Amemiya et al., 1998. Zebrafish hox clusters and vertebrate genome evolution. Science 282: 1711–1714. - PubMed
-
- Aparicio, S., J. Chapman, E. Stupka, N. Putnam, J. M. Chia et al., 2002. Whole-genome shotgun assembly and analysis of the genome of Fugu rubripes. Science 297: 1301–1310. - PubMed
-
- Asahida, T., T. Kobayashi, K. Saitoh and I. Nakayama, 1996. Tissue preservation and total DNA extraction from fish stored at ambient temperature using buffers containing high concentration of urea. Fish. Sci. 62: 727–730.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

