Short fimbriae of Porphyromonas gingivalis and their role in coadhesion with Streptococcus gordonii
- PMID: 15972485
- PMCID: PMC1168573
- DOI: 10.1128/IAI.73.7.3983-3989.2005
Short fimbriae of Porphyromonas gingivalis and their role in coadhesion with Streptococcus gordonii
Abstract
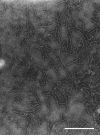
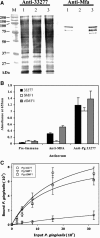
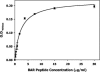
Porphyromonas gingivalis, one of the causative agents of adult periodontitis, attaches and forms biofilms on substrata of Streptococcus gordonii. Coadhesion and biofilm development between these organisms requires the interaction of the short fimbriae of P. gingivalis with the SspB streptococcal surface polypeptide. In this study we investigated the structure and binding activities of the short fimbriae of P. gingivalis. Electron microscopy showed that isolated short fimbriae have an average length of 103 nm and exhibit a helical structure with a pitch of ca. 27 nm. Mfa1, the major protein subunit of the short fimbriae, bound to SspB protein, and this reaction was inhibited by purified recombinant Mfa1 and monospecifc anti-Mfa1 serum in a dose-dependent manner. Complementation of a polar Mfa1 mutant with the mfa1 gene restored the coadhesion phenotype of P. gingivalis. Hence, the Mfa1 structural fimbrial subunit does not require accessory proteins for binding to SspB. Furthermore, the interaction of Mfa1 with SspB is necessary for optimal coadhesion between P. gingivalis and S. gordonii.
Figures

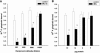
Similar articles
-
Role of the Streptococcus gordonii SspB protein in the development of Porphyromonas gingivalis biofilms on streptococcal substrates.Microbiology (Reading). 2002 Jun;148(Pt 6):1627-1636. doi: 10.1099/00221287-148-6-1627. Microbiology (Reading). 2002. PMID: 12055284
-
Expression of the short fimbriae of Porphyromonas gingivalis is regulated in oral bacterial consortia.FEMS Microbiol Lett. 2006 Sep;262(1):65-71. doi: 10.1111/j.1574-6968.2006.00357.x. FEMS Microbiol Lett. 2006. PMID: 16907740
-
Localization and function of the accessory protein Mfa3 in Porphyromonas gingivalis Mfa1 fimbriae.Mol Oral Microbiol. 2013 Dec;28(6):467-80. doi: 10.1111/omi.12040. Epub 2013 Oct 5. Mol Oral Microbiol. 2013. PMID: 24118823
-
Molecular interaction of Porphyromonas gingivalis with host cells: implication for the microbial pathogenesis of periodontal disease.J Periodontol. 2003 Jan;74(1):90-6. doi: 10.1902/jop.2003.74.1.90. J Periodontol. 2003. PMID: 12593602 Review.
-
In vitro models that support adhesion specificity in biofilms of oral bacteria.Adv Dent Res. 1997 Apr;11(1):33-42. doi: 10.1177/08959374970110011401. Adv Dent Res. 1997. PMID: 9524440 Review.
Cited by
-
Negative correlation of distributions of Streptococcus cristatus and Porphyromonas gingivalis in subgingival plaque.J Clin Microbiol. 2009 Dec;47(12):3902-6. doi: 10.1128/JCM.00072-09. Epub 2009 Oct 21. J Clin Microbiol. 2009. PMID: 19846640 Free PMC article.
-
PgRsp Is a Novel Redox-Sensing Transcription Regulator Essential for Porphyromonas gingivalis Virulence.Microorganisms. 2019 Nov 28;7(12):623. doi: 10.3390/microorganisms7120623. Microorganisms. 2019. PMID: 31795139 Free PMC article.
-
The Application of Small Molecules to the Control of Typical Species Associated With Oral Infectious Diseases.Front Cell Infect Microbiol. 2022 Feb 21;12:816386. doi: 10.3389/fcimb.2022.816386. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 35265531 Free PMC article. Review.
-
Global Noncoding microRNA Profiling in Mice Infected with Partial Human Mouth Microbes (PAHMM) Using an Ecological Time-Sequential Polybacterial Periodontal Infection (ETSPPI) Model Reveals Sex-Specific Differential microRNA Expression.Int J Mol Sci. 2022 May 4;23(9):5107. doi: 10.3390/ijms23095107. Int J Mol Sci. 2022. PMID: 35563501 Free PMC article.
-
Porphyromonas gingivalis HmuY and Streptococcus gordonii GAPDH-Novel Heme Acquisition Strategy in the Oral Microbiome.Int J Mol Sci. 2020 Jun 10;21(11):4150. doi: 10.3390/ijms21114150. Int J Mol Sci. 2020. PMID: 32532033 Free PMC article.
References
-
- Arai, M., N. Hamada, and T. Umemoto. 2000. Purification and characterization of a novel secondary fimbrial protein from Porphyromonas gingivalis strain 381. FEMS Microbiol. Lett. 193:75-81. - PubMed
-
- Chen, W., K. E. Laidig, Y. Park, K. Park, J. R. Yates III, R. J. Lamont, and M. Hackett. 2001. Searching the Porphyromonas gingivalis genome with peptide fragmentation mass spectra. Analyst 126:52-57. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

