Identification of a novel adhesion molecule involved in the virulence of Legionella pneumophila
- PMID: 15972519
- PMCID: PMC1168565
- DOI: 10.1128/IAI.73.7.4272-4280.2005
Identification of a novel adhesion molecule involved in the virulence of Legionella pneumophila
Abstract
Legionella pneumophila is an intracellular bacterium, and its successful parasitism in host cells involves two reciprocal phases: transmission and intracellular replication. In this study, we sought genes that are involved in virulence by screening a genomic DNA library of an L. pneumophila strain, 80-045, with convalescent-phase sera of Legionnaires' disease patients. Three antigens that reacted exclusively with the convalescent-phase sera were isolated. One of them, which shared homology with an integrin analogue of Saccharomyces cerevisiae, was named L. pneumophila adhesion molecule homologous with integrin analogue of S. cerevisiae (LaiA). The laiA gene product was involved in L. pneumophila adhesion to and invasion of the human lung alveolar epithelial cell line A549 during in vitro coculture. However, its presence did not affect multiplication of L. pneumophila within a U937 human macrophage cell line. Furthermore, after intranasal infection of A/J mice, the laiA mutant was eliminated from lungs and caused reduced mortality compared to the wild isolate. Thus, we conclude that the laiA gene encodes a virulence factor that is involved in transmission of L. pneumophila 80-045 and may play a role in Legionnaires' disease in humans.
Figures



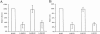
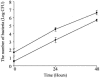

Similar articles
-
bdhA-patD operon as a virulence determinant, revealed by a novel large-scale approach for identification of Legionella pneumophila mutants defective for amoeba infection.Appl Environ Microbiol. 2009 Jul;75(13):4506-15. doi: 10.1128/AEM.00187-09. Epub 2009 May 1. Appl Environ Microbiol. 2009. PMID: 19411431 Free PMC article.
-
Legionella pneumophila entry gene rtxA is involved in virulence.Infect Immun. 2001 Jan;69(1):508-17. doi: 10.1128/IAI.69.1.508-517.2001. Infect Immun. 2001. PMID: 11119544 Free PMC article.
-
Role for the Ankyrin eukaryotic-like genes of Legionella pneumophila in parasitism of protozoan hosts and human macrophages.Environ Microbiol. 2008 Jun;10(6):1460-74. doi: 10.1111/j.1462-2920.2007.01560.x. Epub 2008 Feb 14. Environ Microbiol. 2008. PMID: 18279343
-
Speculations on the influence of infecting phenotype on virulence and antibiotic susceptibility of Legionella pneumophila.J Antimicrob Chemother. 1995 Jul;36(1):7-21. doi: 10.1093/jac/36.1.7. J Antimicrob Chemother. 1995. PMID: 8537286 Review.
-
Analysis of virulence factors of Legionella pneumophila.Zentralbl Bakteriol. 1993 Apr;278(2-3):348-58. doi: 10.1016/s0934-8840(11)80851-0. Zentralbl Bakteriol. 1993. PMID: 8347938 Review.
Cited by
-
Structure of the Legionella Effector, lpg1496, Suggests a Role in Nucleotide Metabolism.J Biol Chem. 2015 Oct 9;290(41):24727-37. doi: 10.1074/jbc.M115.671263. Epub 2015 Aug 20. J Biol Chem. 2015. PMID: 26294765 Free PMC article.
-
The Role of Legionella pneumophila Serogroup 1 Lipopolysaccharide in Host-Pathogen Interaction.Front Microbiol. 2019 Dec 17;10:2890. doi: 10.3389/fmicb.2019.02890. eCollection 2019. Front Microbiol. 2019. PMID: 31921066 Free PMC article.
-
Galactose-1-phosphate uridyltransferase (GalT), an in vivo-induced antigen of Actinobacillus pleuropneumoniae serovar 5b strain L20, provided immunoprotection against serovar 1 strain MS71.PLoS One. 2018 Jun 1;13(6):e0198207. doi: 10.1371/journal.pone.0198207. eCollection 2018. PLoS One. 2018. PMID: 29856812 Free PMC article.
-
Lcl of Legionella pneumophila is an immunogenic GAG binding adhesin that promotes interactions with lung epithelial cells and plays a crucial role in biofilm formation.Infect Immun. 2011 Jun;79(6):2168-81. doi: 10.1128/IAI.01304-10. Epub 2011 Mar 21. Infect Immun. 2011. PMID: 21422183 Free PMC article.
-
Purification of Legiobactin and importance of this siderophore in lung infection by Legionella pneumophila.Infect Immun. 2009 Jul;77(7):2887-95. doi: 10.1128/IAI.00087-09. Epub 2009 Apr 27. Infect Immun. 2009. PMID: 19398549 Free PMC article.
References
-
- Bairoch, A. 1989. PROSITE: a dictionary of protein sites and patterns. University of Geneva, Geneva, Switzerland.
-
- Chiang, S. L., J. J. Mekalanos, and D. W. Holden. 1999. In vivo genetic analysis of bacterial virulence. Annu. Rev. Microbiol. 53:129-154. - PubMed
-
- Chien, M., I. Morozova, S. Shi, H. Sheng, J. Chen, S. M. Gomez, G. Asamani, K. Hill, J. Nuara, M. Feder, J. Rineer, J. J. Greenberg, V. Steshenko, S. H. Park, B. Zhao, E. Teplitskaya, J. R. Edwards, S. Pampou, A. Georghiou, I. C. Chou, W. Iannuccilli, M. E. Ulz, D. H. Kim, A. Geringer-Sameth, C. Goldsberry, P. Morozov, S. G. Fischer, G. Segal, X. Qu, A. Rzhetsky, P. Zhang, E. Cayanis, P. J. De Jong, J. Ju, S. Kalachikov, H. A. Shuman, and J. J. Russo. 2004. The genomic sequence of the accidental pathogen Legionella pneumophila. Science 24:1966-1968. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

