Nonlinear protein degradation and the function of genetic circuits
- PMID: 15972813
- PMCID: PMC1172234
- DOI: 10.1073/pnas.0409553102
Nonlinear protein degradation and the function of genetic circuits
Abstract
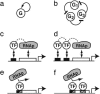
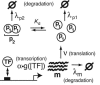
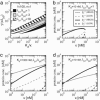
The functions of most genetic circuits require a sufficient degree of cooperativity in the circuit components. Although mechanisms of cooperativity have been studied most extensively in the context of transcriptional initiation control, cooperativity from other processes involved in the operation of the circuits can also play important roles. In this work, we examine a simple kinetic source of cooperativity stemming from the nonlinear degradation of multimeric proteins. Ample experimental evidence suggests that protein subunits can degrade less rapidly when associated in multimeric complexes, an effect we refer to as "cooperative stability." For dimeric transcription factors, this effect leads to a concentration-dependence in the degradation rate because monomers, which are predominant at low concentrations, will be more rapidly degraded. Thus, cooperative stability can effectively widen the accessible range of protein levels in vivo. Through theoretical analysis of two exemplary genetic circuits in bacteria, we show that such an increased range is important for the robust operation of genetic circuits as well as their evolvability. Our calculations demonstrate that a few-fold difference between the degradation rate of monomers and dimers can already enhance the function of these circuits substantially. We discuss molecular mechanisms of cooperative stability and their occurrence in natural or engineered systems. Our results suggest that cooperative stability needs to be considered explicitly and characterized quantitatively in any systematic experimental or theoretical study of gene circuits.
Figures
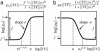
References
-
- Jenal, U. & Hengge-Aronis, R. (2003) Curr. Opin. Microbiol. 6 163-172. - PubMed
-
- Gonzalez, M. & Woodgate, R. (2002) BioEssays 24 141-148. - PubMed
-
- Johnson, P. R., Swanson, R., Rakhilina, L. & Hochstrasser, M. (1998) Cell 94 217-227. - PubMed
-
- Parsell, D. A. & Sauer, R. T. (1989) J. Biol. Chem. 264 7590-7595. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

