Generation of protein kinase Ck1alpha mutants which discriminate between canonical and non-canonical substrates
- PMID: 15975091
- PMCID: PMC1276941
- DOI: 10.1042/BJ20050717
Generation of protein kinase Ck1alpha mutants which discriminate between canonical and non-canonical substrates
Abstract
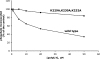
Protein kinase CK1 denotes a family of pleiotropic serine/threonine protein kinases implicated in a variety of cellular functions. Typically, CK1 acts as a 'phosphate-directed' kinase whose targeting is primed by a single phosphorylated side chain at position n-3 or n-4 relative to serine/threonine, but increasing evidence is accumulating that CK1 can also engage some of its substrates at sites that do not conform to this canonical consensus. In the present paper, we show that CK1a phosphorylates with the same efficiency phosphopeptides primed by a phosphoserine residue at either n-3 [pS(-3)] or n-4 [pS(-4)] positions. The phosphorylation efficiency of the pS(-4) peptide, and to a lesser extent that of the pS(-3) peptide, is impaired by the triple mutation of the lysine residues in the K229KQK232 stretch to alanine residues, promoting 40-fold and 6-fold increases of Km respectively. In both cases, the individual mutation of Lys232 is as detrimental as the triple mutation. A kinetic alanine-scan analysis with a series of substituted peptide substrates in which the priming phosphoserine residue was effectively replaced by a cluster of four aspartate residues was also consistent with a crucial role of Lys232 in the recognition of the acidic determinant at position n-4. In sharp contrast, the phosphorylation of b-catenin and of a peptide including the non-canonical b-catenin site (Ser45) lacking acidic/phosphorylated determinants upstream is not significantly affected by mutations in the KKQK stretch. These data provide a molecular insight into the structural features that underlie the site specificity of CK1a and disclose the possibility of developing strategies for the preferential targeting of subsets of CK1 substrates.
Figures

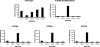
References
-
- Manning G., Whyte D. B., Martinez R., Hunter T., Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. - PubMed
-
- Knippschild U., Gocht A., Wolff S., Huber N., Loehler J., Stoeter M. The casein kinase 1 family: participation in multiple cellular processes in eukaryotes. Cell. Signalling. 2005;17:675–689. - PubMed
-
- Vielhaber E., Virshup D. M. Casein kinase I: from obscurity to center stage. Life. 2001;51:73–78. - PubMed
-
- Gross S., Anderson R. Casein kinase I: spatial organization and positioning of a multifunctional protein kinase family. Cell. Signalling. 1998;10:699–711. - PubMed
-
- Hoekstra M. F., Liskay R. M., Ou A. C., DeMaggio A. J., Burbee D. G., Heffron F. HRR25, a putative protein kinase from budding yeast: association with repair of damaged DNA. Science. 1991;253:1031–1034. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

