The novel fission yeast protein Pal1p interacts with Hip1-related Sla2p/End4p and is involved in cellular morphogenesis
- PMID: 15975911
- PMCID: PMC1196324
- DOI: 10.1091/mbc.e04-11-0976
The novel fission yeast protein Pal1p interacts with Hip1-related Sla2p/End4p and is involved in cellular morphogenesis
Abstract
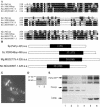
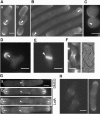
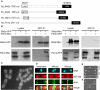
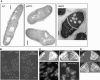
The establishment and maintenance of characteristic cellular morphologies is a fundamental property of all cells. Here we describe Schizosaccharomyces pombe Pal1p, a protein important for maintenance of cylindrical cellular morphology. Pal1p is a novel membrane-associated protein that localizes to the growing tips of interphase cells and to the division site in cells undergoing cytokinesis in an F-actin- and microtubule-independent manner. Cells deleted for pal1 display morphological defects, characterized by the occurrence of spherical and pear-shaped cells with an abnormal cell wall. Pal1p physically interacts and displays overlapping localization with the Huntingtin-interacting-protein (Hip1)-related protein Sla2p/End4p, which is also required for establishment of cylindrical cellular morphology. Sla2p is important for efficient localization of Pal1p to the sites of polarized growth and appears to function upstream of Pal1p. Interestingly, spherical pal1Delta mutants polarize to establish a pearlike morphology before mitosis in a manner dependent on the kelch-repeat protein Tea1p and the cell cycle inhibitory kinase Wee1p. Thus, overlapping mechanisms involving Pal1p, Tea1p, and Sla2p contribute to the establishment of cylindrical cellular morphology, which is important for proper spatial regulation of cytokinesis.
Figures


Similar articles
-
Role of bud6p and tea1p in the interaction between actin and microtubules for the establishment of cell polarity in fission yeast.Curr Biol. 2001 Jun 5;11(11):836-45. doi: 10.1016/s0960-9822(01)00235-4. Curr Biol. 2001. PMID: 11516644
-
Regulation of a formin complex by the microtubule plus end protein tea1p.J Cell Biol. 2004 Jun 7;165(5):697-707. doi: 10.1083/jcb.200403090. J Cell Biol. 2004. PMID: 15184402 Free PMC article.
-
Regulation of actin assembly by microtubules in fission yeast cell polarity.Novartis Found Symp. 2005;269:59-66; discussion 66-72, 223-30. Novartis Found Symp. 2005. PMID: 16355535
-
Role of microtubules and tea1p in establishment and maintenance of fission yeast cell polarity.J Cell Sci. 2004 Feb 15;117(Pt 5):689-700. doi: 10.1242/jcs.00925. Epub 2004 Jan 20. J Cell Sci. 2004. PMID: 14734657
-
Microtubule-dependent cell morphogenesis in the fission yeast.Trends Cell Biol. 2009 Sep;19(9):447-54. doi: 10.1016/j.tcb.2009.06.003. Epub 2009 Aug 25. Trends Cell Biol. 2009. PMID: 19713114 Review.
Cited by
-
Regulation of fission yeast morphogenesis by PP2A activator pta2.PLoS One. 2012;7(3):e32823. doi: 10.1371/journal.pone.0032823. Epub 2012 Mar 5. PLoS One. 2012. PMID: 22403715 Free PMC article.
-
Mechanisms Connecting the Conserved Protein Kinases Ssp1, Kin1, and Pom1 in Fission Yeast Cell Polarity and Division.Curr Biol. 2018 Jan 8;28(1):84-92.e4. doi: 10.1016/j.cub.2017.11.034. Epub 2017 Dec 14. Curr Biol. 2018. PMID: 29249658 Free PMC article.
-
Actin and endocytosis in budding yeast.Genetics. 2015 Feb;199(2):315-58. doi: 10.1534/genetics.112.145540. Genetics. 2015. PMID: 25657349 Free PMC article. Review.
-
A genome-wide screening of potential target genes to enhance the antifungal activity of micafungin in Schizosaccharomyces pombe.PLoS One. 2013 May 30;8(5):e65904. doi: 10.1371/journal.pone.0065904. Print 2013. PLoS One. 2013. PMID: 23738021 Free PMC article.
-
A tudor domain protein SPINDLIN1 interacts with the mRNA-binding protein SERBP1 and is involved in mouse oocyte meiotic resumption.PLoS One. 2013 Jul 22;8(7):e69764. doi: 10.1371/journal.pone.0069764. Print 2013. PLoS One. 2013. PMID: 23894536 Free PMC article.
References
-
- Arellano, M., Niccoli, T., and Nurse, P. (2002). Tea3p is a cell end marker activating polarized growth in Schizosaccharomyces pombe. Curr. Biol. 12, 751-756. - PubMed
-
- Balasubramanian, M. K., Helfman, D. M., and Hemmingsen, S. M. (1992). A new tropomyosin essential for cytokinesis in the fission yeast S. pombe. Nature 360, 84-87. - PubMed
-
- Balasubramanian, M. K., McCollum, D., and Gould, K. L. (1997). Cytokinesis in fission yeast Schizosaccharomyces pombe. Methods Enzymol. 283, 494-506. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

