Structural and biochemical basis for selective repression of the orphan nuclear receptor liver receptor homolog 1 by small heterodimer partner
- PMID: 15976031
- PMCID: PMC1157103
- DOI: 10.1073/pnas.0501204102
Structural and biochemical basis for selective repression of the orphan nuclear receptor liver receptor homolog 1 by small heterodimer partner
Abstract
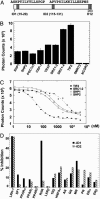
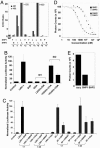
The functional interaction between the orphan nuclear receptors small heterodimer partner (SHP) and liver receptor homolog 1 (LRH-1), where SHP binds to LRH-1 and represses its constitutive transcriptional activity, is crucial for regulating genes involved in cholesterol homeostasis. Here, we report structural and biochemical analyses of the LRH-1/SHP interaction. The crystal structure and modeling studies of the LRH-1 ligand-binding domain bound to either of the two LXXLL-related motifs of SHP show that the receptor undergoes conformational changes to accommodate the SHP docking and reveal key residues that determine the potency and selectivity of SHP binding. Through a combination of mutagenesis and binding studies, we demonstrate that only the second SHP LXXLL motif is required for repressing LRH-1, and this motif displays a strong preference for binding to LRH-1 over the closely related receptor steroidogeneic factor 1 (SF-1). Structural comparisons indicate that this binding selectivity is determined by residues flanking the core LXXLL motifs. These results establish a structural model for understanding how SHP interacts with LRH-1 to regulate cholesterol homeostasis and provide new insights into how nuclear receptor/coregulator selectivity is achieved.
Figures



Similar articles
-
Dual mechanisms for repression of the monomeric orphan receptor liver receptor homologous protein-1 by the orphan small heterodimer partner.J Biol Chem. 2002 Jan 25;277(4):2463-7. doi: 10.1074/jbc.M105161200. Epub 2001 Oct 19. J Biol Chem. 2002. PMID: 11668176
-
Ligand-dependent regulation of the activity of the orphan nuclear receptor, small heterodimer partner (SHP), in the repression of bile acid biosynthetic CYP7A1 and CYP8B1 genes.Mol Endocrinol. 2011 Jul;25(7):1159-69. doi: 10.1210/me.2011-0033. Epub 2011 May 12. Mol Endocrinol. 2011. PMID: 21566081 Free PMC article.
-
Genome-wide analysis of LXXLL-mediated DAX1/SHP-nuclear receptor interaction network and rational design of stapled LXXLL-based peptides to target the specific network profile.Int J Biol Macromol. 2019 May 15;129:13-22. doi: 10.1016/j.ijbiomac.2019.02.014. Epub 2019 Feb 4. Int J Biol Macromol. 2019. PMID: 30731167
-
Ligand-independent actions of the orphan receptors/corepressors DAX-1 and SHP in metabolism, reproduction and disease.J Steroid Biochem Mol Biol. 2012 Jul;130(3-5):169-79. doi: 10.1016/j.jsbmb.2011.04.007. Epub 2011 Apr 29. J Steroid Biochem Mol Biol. 2012. PMID: 21550402 Review.
-
LRH-1: an orphan nuclear receptor involved in development, metabolism and steroidogenesis.Trends Cell Biol. 2004 May;14(5):250-60. doi: 10.1016/j.tcb.2004.03.008. Trends Cell Biol. 2004. PMID: 15130581 Review.
Cited by
-
Novel Thiazolidinedione and Rhodanine Derivatives Regulate Glucose Metabolism, Improve Insulin Sensitivity, and Activate the Peroxisome Proliferator-Activated γ Receptor.ACS Omega. 2024 Jan 25;9(5):5463-5484. doi: 10.1021/acsomega.3c07149. eCollection 2024 Feb 6. ACS Omega. 2024. PMID: 38343951 Free PMC article.
-
Novel polymorphisms of nuclear receptor SHP associated with functional and structural changes.J Biol Chem. 2010 Aug 6;285(32):24871-81. doi: 10.1074/jbc.M110.133280. Epub 2010 Jun 1. J Biol Chem. 2010. PMID: 20516075 Free PMC article.
-
Insights into Orphan Nuclear Receptors as Prognostic Markers and Novel Therapeutic Targets for Breast Cancer.Front Endocrinol (Lausanne). 2015 Aug 7;6:115. doi: 10.3389/fendo.2015.00115. eCollection 2015. Front Endocrinol (Lausanne). 2015. PMID: 26300846 Free PMC article. Review.
-
Structural overview and perspectives of the nuclear receptors, a major family as the direct targets for small-molecule drugs.Acta Biochim Biophys Sin (Shanghai). 2022 Jan 25;54(1):12-24. doi: 10.3724/abbs.2021001. Acta Biochim Biophys Sin (Shanghai). 2022. PMID: 35130630 Free PMC article.
-
Virtual Screening as a Technique for PPAR Modulator Discovery.PPAR Res. 2010;2010:861238. doi: 10.1155/2010/861238. Epub 2009 Sep 2. PPAR Res. 2010. PMID: 19746174 Free PMC article.
References
-
- Fayard, E., Auwerx, J. & Schoonjans, K. (2004) Trends Cell Biol. 14, 250-260. - PubMed
-
- Goodwin, B., Jones, S. A., Price, R. R., Watson, M. A., McKee, D. D., Moore, L. B., Galardi, C., Wilson, J. G., Lewis, M. C., Roth, M. E., et al. (2000) Mol. Cell 6, 517-526. - PubMed
-
- Lu, T. T., Makishima, M., Repa, J. J., Schoonjans, K., Kerr, T. A., Auwerx, J. & Mangelsdorf, D. J. (2000) Mol. Cell 6, 507-515. - PubMed
-
- Delerive, P., Galardi, C. M., Bisi, J. E., Nicodeme, E. & Goodwin, B. (2004) Mol. Endocrinol. 18, 2378-2387. - PubMed
-
- Chen, F., Ma, L., Dawson, P. A., Sinal, C. J., Sehayek, E., Gonzalez, F. J., Breslow, J., Ananthanarayanan, M. & Shneider, B. L. (2003) J. Biol. Chem. 278, 19909-19916. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

