Dissecting defense-related and developmental transcriptional responses of maize during Ustilago maydis infection and subsequent tumor formation
- PMID: 15980197
- PMCID: PMC1176445
- DOI: 10.1104/pp.105.061200
Dissecting defense-related and developmental transcriptional responses of maize during Ustilago maydis infection and subsequent tumor formation
Abstract
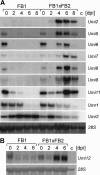
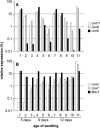
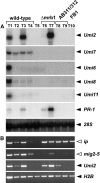
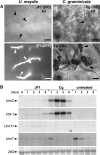
Infection of maize (Zea mays) plants with the smut fungus Ustilago maydis triggers the formation of tumors on aerial parts in which the fungal life cycle is completed. A differential display screen was performed to gain insight into transcriptional changes of the host response. Some of the genes strongly up-regulated in tumors showed a pronounced developmental expression pattern with decreasing transcript levels from basal to apical shoot segments, suggesting that U. maydis has the capacity to extend the undifferentiated state of maize plants. Differentially expressed genes implicated in secondary metabolism were Bx1, involved in biosynthesis of the cyclic hydroxamic acid 2,4-dihydroxy-7-methoxy-2H-1,4-benzoxazin-3-one, and a novel putative sesquiterpene cyclase gene U. maydis induced (Umi)2. Together with the up-regulation of Umi11 encoding a cyclotide-like protein this suggests a nonconventional induction of plant defenses. Explicitly, U. maydis was resistant to 2,4-dihydroxy-7-methoxy-2H-1,4-benzoxazin-3-one but susceptible to its benzoxazolinone derivative 6-methoxy-2-benzoxazolinone. Infection studies of isolated leaves with U. maydis and Colletotrichum graminicola provided evidence for coregulation of Umi2 and PR-1 gene expression, with mRNA levels strongly determined by the extent of fungal colonization within tissue. However, in contrast to Umi2, transcript levels of PR-1 remained low in plants infected with wild-type U. maydis but were 8-fold elevated upon infection with an U. maydis mutant strongly attenuated in pathogenic development. This suggests that U. maydis colonization in planta suppresses a classical defense response. Furthermore, comparative expression analysis uncovered distinct transcriptional programs operating in the host in response to fungal infection and subsequent tumor formation.
Figures



References
-
- Banuett F, Herskowitz I (1996) Discrete developmental stages during teliospore formation in the corn smut fungus, Ustilago maydis. Development 122: 2965–2976 - PubMed
-
- Basse CW, Kolb S, Kahmann R (2002. a) A maize-specifically expressed gene cluster in Ustilago maydis. Mol Microbiol 43: 75–93 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

