D-GLYCERATE 3-KINASE, the last unknown enzyme in the photorespiratory cycle in Arabidopsis, belongs to a novel kinase family
- PMID: 15980259
- PMCID: PMC1182498
- DOI: 10.1105/tpc.105.033993
D-GLYCERATE 3-KINASE, the last unknown enzyme in the photorespiratory cycle in Arabidopsis, belongs to a novel kinase family
Abstract
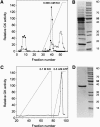
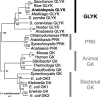
D-GLYCERATE 3-KINASE (GLYK; EC 2.7.1.31) catalyzes the concluding reaction of the photorespiratory C2 cycle, an indispensable ancillary metabolic pathway to the photosynthetic C3 cycle that enables land plants to grow in an oxygen-containing atmosphere. Except for GLYK, all other enzymes that contribute to the C2 cycle are known by their primary structures, and the encoding genes have been identified. We have purified and partially sequenced this yet missing enzyme from Arabidopsis thaliana and identified it as a putative kinase-annotated single-copy gene At1g80380. The exclusive catalytic properties of the gene product were confirmed after heterologous expression in Escherichia coli. Arabidopsis T-DNA insertional knockout mutants show no GLYK activity and are not viable in normal air; however, they grow under elevated CO2, providing direct evidence of the obligatory nature of the ultimate step of the C2 cycle. The newly identified GLYK is both structurally and phylogenetically distinct from known glycerate kinases from bacteria and animals. Orthologous enzymes are present in other plants, fungi, and some cyanobacteria. The metabolic context of GLYK activity in fungi and cyanobacteria remains to be investigated.
Figures




References
-
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. - PubMed
-
- Arabidopsis Genome Initiative (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408 796–815. - PubMed
-
- Black, S., and Wright, N.G. (1956). Enzymatic formation of glyceryl and phosphoglyceryl methylthiol esters. J. Biol. Chem. 221 171–180. - PubMed
-
- Blackwell, R.D., Murray, A.J.S., Lea, P.J., Kendall, A., Hall, N.P., Turner, J.C., and Wallsgrove, R.M. (1988). The value of mutants unable to carry out photorespiration. Photosynth. Res. 16 155–176. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

