Identification of novel genes in Arabidopsis involved in secondary cell wall formation using expression profiling and reverse genetics
- PMID: 15980264
- PMCID: PMC1182489
- DOI: 10.1105/tpc.105.031542
Identification of novel genes in Arabidopsis involved in secondary cell wall formation using expression profiling and reverse genetics
Abstract
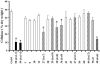
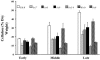
Forward genetic screens have led to the isolation of several genes involved in secondary cell wall formation. A variety of evidence, however, suggests that the list of genes identified is not exhaustive. To address this problem, microarray data have been generated from tissue undergoing secondary cell wall formation and used to identify genes that exhibit a similar expression pattern to the secondary cell wall-specific cellulose synthase genes IRREGULAR XYLEM1 (IRX1) and IRX3. Cross-referencing this analysis with publicly available microarray data resulted in the selection of 16 genes for reverse genetic analysis. Lines containing an insertion in seven of these genes exhibited a clear irx phenotype characteristic of a secondary cell wall defect. Only one line, containing an insertion in a member of the COBRA gene family, exhibited a large decrease in cellulose content. Five of the genes identified as being essential for secondary cell wall biosynthesis have not been previously characterized. These genes are likely to define entirely novel processes in secondary cell wall formation and illustrate the success of combining expression data with reverse genetics to address gene function.
Figures








References
-
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. - PubMed
-
- Arabidopsis Genome Initiative (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408 796–815. - PubMed
-
- Birnbaum, K., Shasha, D.E., Wang, J.Y., Jung, J.W., Lambert, G.M., Galbraith, D.W., and Benfey, P.N. (2003). A gene expression map of the Arabidopsis root. Science 302 1956–1960. - PubMed
-
- Bolstad, B.M., Irizarry, R.A., Astrand, M., and Speed, T.P. (2003). A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19 185–193. - PubMed
-
- Bouton, S., Leboeuf, E., Mouille, G., Leydecker, M.T., Talbotec, J., Granier, F., Lahaye, M., Hofte, H., and Truong, H.N. (2002). Quasimodo1 encodes a putative membrane-bound glycosyltransferase required for normal pectin synthesis and cell adhesion in Arabidopsis. Plant Cell 14 2577–2590. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

