Multiple-antibiotic resistance in Salmonella enterica serotype Paratyphi B isolates collected in France between 2000 and 2003 is due mainly to strains harboring Salmonella genomic islands 1, 1-B, and 1-C
- PMID: 15980351
- PMCID: PMC1168691
- DOI: 10.1128/AAC.49.7.2793-2801.2005
Multiple-antibiotic resistance in Salmonella enterica serotype Paratyphi B isolates collected in France between 2000 and 2003 is due mainly to strains harboring Salmonella genomic islands 1, 1-B, and 1-C
Abstract
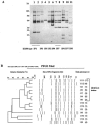
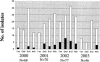
This study was conducted to investigate the occurrence of multiple-antibiotic resistance among 261 clinical isolates of Salmonella enterica serotype Paratyphi B strains collected between 2000 and 2003 through the network of the French National Reference Center for Salmonella. The 47 multidrug-resistant (MDR) isolates identified (18%), were characterized on the basis of the presence of several resistance genes (bla(TEM), bla(PSE-1), bla(CTX-M), floR, aadA2, qacEdelta1, and sul1), the presence of Salmonella genomic island 1 (SGI1) by PCR mapping and hybridization, and the clonality of these isolates by several molecular (ribotyping, IS200 profiling, and pulsed-field gel electrophoresis [PFGE]) and phage typing methods. The results of PCR and Southern blot experiments indicated that 39 (83%) of the 47 S. enterica serotype Paratyphi B biotype Java MDR isolates possessed the SGI1 cluster (MDR/SGI1). Among these 39 MDR/SGI1 isolates, only 3 contained variations in SGI1, SGI1-B (n = 1) and SGI1-C (n = 2). The 39 MDR/SGI1 isolates showed the same specific PstI-IS200 profile 1, which contained seven copies from 2.6 to 18 kb. Two PstI ribotypes were found in MDR/SGI1 isolates, RP1 (n = 38) and RP6 (n = 1). Ribotype RP1 was also found in two susceptible strains. Analysis by PFGE using XbaI revealed that all the MDR/SGI1 isolates were grouped in two related clusters, with a similarity percentage of 82%. Isolation of MDR/SGI1 isolates in France was observed mainly between the second quarter of 2001 and the end of 2002. The source of the contamination has not been identified to date. A single isolate possessing the extended-spectrum beta-lactamase bla(CTX-M-15) gene was also identified during the study.
Figures

References
-
- Boyd, D., G. A. Peters, A. Cloeckaert, K. S. Boumedine, E. Chaslus-Dancla, H. Imberechts, and M. R. Mulvey. 2001. Complete nucleotide sequence of a 43-kilobase genomic island associated with the multidrug resistance region of Salmonella enterica serovar Typhimurium DT104 and its identification in phage type DT120 and serovar Agona. J. Bacteriol. 183:5725-5732. - PMC - PubMed
-
- Brown, D. J., H. Mather, L. M. Browning, and J. E. Coia. 2003. Investigation of human infections with Salmonella enterica serovar Java in Scotland and possible association with imported poultry. Euro. Surveill. 8:35-40. - PubMed
-
- Desenclos, J. C., P. Bouvet, E. Benz-Lemoine, F. Grimont, H. Desqueyroux, I. Rebiere, and P. A. D. Grimont. 1996. Large outbreak of Salmonella enterica serotype paratyphi B infection caused by a goats' milk cheese, France, 1993: a case finding and epidemiological study. BMJ 312:91-94. - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

